Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas - PubMed (original) (raw)
Meta-Analysis
. 2012 Apr;123(4):473-84.
doi: 10.1007/s00401-012-0958-8. Epub 2012 Feb 23.
Andrey Korshunov, Marc Remke, David T W Jones, Maria Schlanstein, Paul A Northcott, Yoon-Jae Cho, Jan Koster, Antoinette Schouten-van Meeteren, Dannis van Vuurden, Steven C Clifford, Torsten Pietsch, Andre O von Bueren, Stefan Rutkowski, Martin McCabe, V Peter Collins, Magnus L Bäcklund, Christine Haberler, Franck Bourdeaut, Olivier Delattre, Francois Doz, David W Ellison, Richard J Gilbertson, Scott L Pomeroy, Michael D Taylor, Peter Lichter, Stefan M Pfister
Affiliations
- PMID: 22358457
- PMCID: PMC3306778
- DOI: 10.1007/s00401-012-0958-8
Meta-Analysis
Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas
Marcel Kool et al. Acta Neuropathol. 2012 Apr.
Abstract
Medulloblastoma is the most common malignant brain tumor in childhood. Molecular studies from several groups around the world demonstrated that medulloblastoma is not one disease but comprises a collection of distinct molecular subgroups. However, all these studies reported on different numbers of subgroups. The current consensus is that there are only four core subgroups, which should be termed WNT, SHH, Group 3 and Group 4. Based on this, we performed a meta-analysis of all molecular and clinical data of 550 medulloblastomas brought together from seven independent studies. All cases were analyzed by gene expression profiling and for most cases SNP or array-CGH data were available. Data are presented for all medulloblastomas together and for each subgroup separately. For validation purposes, we compared the results of this meta-analysis with another large medulloblastoma cohort (n = 402) for which subgroup information was obtained by immunohistochemistry. Results from both cohorts are highly similar and show how distinct the molecular subtypes are with respect to their transcriptome, DNA copy-number aberrations, demographics, and survival. Results from these analyses will form the basis for prospective multi-center studies and will have an impact on how the different subgroups of medulloblastoma will be treated in the future.
Figures
Fig. 1
Demographic distribution of medulloblastoma subgroups. Subgroup distribution is shown for all medulloblastoma patients (a), infants (age <4 years) (**b**), children (age 4–16) (**c**), and adults (age >16) (d). Numbers on the Y axis indicate number of patients. Male:female frequencies are shown for all four subgroups in all patients (e), infants (f), children (g), and adults (h). Males are indicated in blue, females in pink. Distribution of histological subtype is shown for all four subgroups in all patients (i), infants (j), children (k), and adults (l). Classic histology is indicated in dark red, desmoplastic/extensive nodular histology in gray, and large cell/anaplastic histology in orange. Frequencies of metastasized (green) and non-metastasized (light green) cases are shown for all four subgroups in all patients (m), infants (n), children (o), and adults (p). Age distribution shown for males (solid lines) and females (dotted lines) is plotted for each of the four subgroups: WNT (q), SHH (r), Group 3 (s), and Group 4 (t). Numbers on the Y axis indicate the frequency of that particular subgroup within the indicated age group (in years) on the X axis among all patients
Fig. 2
Overview of chromosomal aberrations in the four medulloblastoma subgroups. Array-CGH and SNP data were scored for loss (green), gain (red), or no change (gray) for all chromosomal arms. Results were plotted as frequencies at which these aberrations occurred within each molecular subgroup. P values on the right indicate whether there was a significant difference in the distribution of these frequencies across the four subgroups (Chi-square test). NS not significant
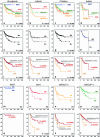
Fig. 3
Overall survival (OS) analyses of molecular, clinical, and histological subgroups within the gene expression profiling cohort using Kaplan–Meier plots and log-rank tests. OS analysis of molecular subgroups among all patients (a), infants (b), children (c), and adults (d). OS analysis of metastasized (M1–M4, indicated as M+) versus non-metastasized (M0) cases, plotted for all patients (e), infants (f), children (g), and adults (h). OS analyses of classic, desmoplastic and LCA histological subgroups among all patients (i), infants (j), children (k), and adults (l). OS analysis of metastasized (M1–M4, indicated as M+) versus non-metastasized (M0) cases, plotted for each molecular subgroup: WNT (m), SHH (n), Group 3 (o), and Group 4 (p). OS analyses of classic, desmoplastic and LCA histological subgroups plotted for each molecular subgroup: WNT (q), SHH (r), Group 3 (s), and Group 4 (t). Numbers on the Y axis indicate the fraction of surviving patients. Numbers on the X axis indicate the follow-up time in months. NS not significant
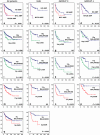
Fig. 4
Overall survival (OS) analyses of cytogenetic subgroups using Kaplan–Meier plots and log-rank tests for statistical significance. Data are shown for all patients within the gene expression profiling cohort, for the SHH subgroup, for Group 3, and for Group 4. OS analysis of patients having a MYC or MYCN amplification versus patients not having these amplifications. e–h OS analysis of patients harboring 10q loss versus patients with a balanced 10q. i–l OS analysis of patients harboring 17p loss versus patients with a balanced 17p. m–p OS analysis of patients harboring 17q gain versus patients with a balanced 17q. q–r OS analysis of patients harboring 3q gain versus patients with a balanced 3q. Frequency of 3q gain within Group 3 and Group 4 was too low to perform survival analyses. Numbers on the Y axis indicate the fraction of surviving patients. Numbers on the X axis indicate the follow-up time in months. NS not significant
Similar articles
- Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.
Elwenspoek MM, Thom H, Sheppard AL, Keeney E, O'Donnell R, Jackson J, Roadevin C, Dawson S, Lane D, Stubbs J, Everitt H, Watson JC, Hay AD, Gillett P, Robins G, Jones HE, Mallett S, Whiting PF. Elwenspoek MM, et al. Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article. - Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis.
Ramaswamy V, Remke M, Bouffet E, Faria CC, Perreault S, Cho YJ, Shih DJ, Luu B, Dubuc AM, Northcott PA, Schüller U, Gururangan S, McLendon R, Bigner D, Fouladi M, Ligon KL, Pomeroy SL, Dunn S, Triscott J, Jabado N, Fontebasso A, Jones DT, Kool M, Karajannis MA, Gardner SL, Zagzag D, Nunes S, Pimentel J, Mora J, Lipp E, Walter AW, Ryzhova M, Zheludkova O, Kumirova E, Alshami J, Croul SE, Rutka JT, Hawkins C, Tabori U, Codispoti KE, Packer RJ, Pfister SM, Korshunov A, Taylor MD. Ramaswamy V, et al. Lancet Oncol. 2013 Nov;14(12):1200-7. doi: 10.1016/S1470-2045(13)70449-2. Epub 2013 Oct 17. Lancet Oncol. 2013. PMID: 24140199 Free PMC article. - Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.
Triana L, Palacios Huatuco RM, Campilgio G, Liscano E. Triana L, et al. Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
- Clinical and Molecular Characteristics and Outcome of Adult Medulloblastoma at a Tertiary Cancer Center.
Almousa A, Erjan A, Sarhan N, Obeidat M, Alshorbaji A, Amarin R, Alawabdeh T, Abu-Hijlih R, Mujlli M, Kh Ibrahimi A, Abu Laban D, Maraqa B, Al-Ani A, Al Sharie S, Al-Hussaini M. Almousa A, et al. Cancers (Basel). 2024 Oct 25;16(21):3609. doi: 10.3390/cancers16213609. Cancers (Basel). 2024. PMID: 39518048 Free PMC article. - Application of radiomics for diagnosis, subtyping, and prognostication of medulloblastomas: a systematic review.
Fotouhi M, Shahbandi A, Mehr FSK, Shahla MM, Nouredini SM, Kankam SB, Khorasanizadeh M, Chambless LB. Fotouhi M, et al. Neurosurg Rev. 2024 Oct 29;47(1):827. doi: 10.1007/s10143-024-03060-1. Neurosurg Rev. 2024. PMID: 39467891 Review. - Differential regulation of Shh-Gli1 cell signalling pathway on homeodomain transcription factors Nkx2.2 and Pax6 during the medulloblastoma genesis.
P M MM, Farheen S, Sharma RM, Shahi MH. P M MM, et al. Mol Biol Rep. 2024 Oct 26;51(1):1096. doi: 10.1007/s11033-024-10026-5. Mol Biol Rep. 2024. PMID: 39460795 - Immunoexpression of Survivin and P53 in the Histological Subtypes of Medulloblastoma: A Cross-Sectional Observational Study.
G G, Bharti S, Jha VC, Nigam JS, Ganesh R A, Bhadani P. G G, et al. Cureus. 2024 Jul 29;16(7):e65627. doi: 10.7759/cureus.65627. eCollection 2024 Jul. Cureus. 2024. PMID: 39205763 Free PMC article. - Radiomics and artificial intelligence applications in pediatric brain tumors.
Pacchiano F, Tortora M, Doneda C, Izzo G, Arrigoni F, Ugga L, Cuocolo R, Parazzini C, Righini A, Brunetti A. Pacchiano F, et al. World J Pediatr. 2024 Aug;20(8):747-763. doi: 10.1007/s12519-024-00823-0. Epub 2024 Jun 27. World J Pediatr. 2024. PMID: 38935233 Free PMC article. Review.
References
- Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, Lau CC, Olson JM, Gilbertson RJ, Gajjar A, Delattre O, Kool M, Ligon K, Meyerson M, Mesirov JP, Pomeroy SL. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29(11):1424–1430. doi: 10.1200/JCO.2010.28.5148. - DOI - PMC - PubMed
- Clifford SC, Lusher ME, Lindsey JC, Langdon JA, Gilbertson RJ, Straughton D, Ellison DW. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5(22):2666–2670. doi: 10.4161/cc.5.22.3446. - DOI - PubMed
- Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, Taylor RE, Bailey S, Clifford SC, Gilbertson RJ. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121(3):381–396. doi: 10.1007/s00401-011-0800-8. - DOI - PMC - PubMed
- Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, Taylor RE, Pearson AD, Clifford SC. Beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23(31):7951–7957. doi: 10.1200/JCO.2005.01.5479. - DOI - PubMed
- Fattet S, Haberler C, Legoix P, Varlet P, Lellouch-Tubiana A, Lair S, Manie E, Raquin MA, Bours D, Carpentier S, Barillot E, Grill J, Doz F, Puget S, Janoueix-Lerosey I, Delattre O. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol. 2009;218(1):86–94. doi: 10.1002/path.2514. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources