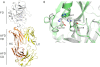Inhibiting alternative pathway complement activation by targeting the factor D exosite - PubMed (original) (raw)
Inhibiting alternative pathway complement activation by targeting the factor D exosite
Kenneth J Katschke Jr et al. J Biol Chem. 2012.
Abstract
By virtue of its amplifying property, the alternative complement pathway has been implicated in a number of inflammatory diseases and constitutes an attractive therapeutic target. An anti-factor D Fab fragment (AFD) was generated to inhibit the alternative complement pathway in advanced dry age-related macular degeneration. AFD potently prevented factor D (FD)-mediated proteolytic activation of its macromolecular substrate C3bB, but not proteolysis of a small synthetic substrate, indicating that AFD did not block access of the substrate to the catalytic site. The crystal structures of AFD in complex with human and cynomolgus FD (at 2.4 and 2.3 Å, respectively) revealed the molecular details of the inhibitory mechanism. The structures show that the AFD-binding site includes surface loops of FD that form part of the FD exosite. Thus, AFD inhibits FD proteolytic function by interfering with macromolecular substrate access rather than by inhibiting FD catalysis, providing the molecular basis of AFD-mediated inhibition of a rate-limiting step in the alternative complement pathway.
Figures
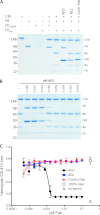
FIGURE 1.
AFD blocks activation of C3bB proconvertase. A, SDS-PAGE of proconvertase formation in the presence of catalytically inactive FD (FD(S195A)) and wild-type FD and the effects of blocking (AFD) or non-blocking (8E2) FD antibodies or anti-hepsin (control) Fab fragments. B, AFD fully inhibited C3 proconvertase activation (indicated by the absence of Ba and Bb formation) at a 1:2 molar ratio of AFD to FD. Convertase formation was measured by conversion of FB to C3Ba and C3Bb. C, AFD blocked hemolysis in an assay selective for the alternative pathway. Maximal blockade of hemolysis was achieved at a 1:1 molar ratio of AFD to FD.
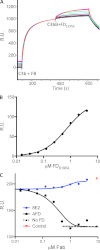
FIGURE 2.
AFD blocks binding of FD to C3bB proconvertase. A, binding of FD to a preformed C3bB complex as shown by surface plasmon resonance. A fixed concentration of FB (1 μ
m
) was injected over immobilized C3b, followed by a dilution series of enzymatically inactive FD (FD(S195A), 2-fold dilution series of 4.0 μ
m
to 15.6 n
m
). B, the binding affinity of FD for C3bB (720 n
m
) was measured by detecting the response (y axis) to increasing concentrations of FD(S195A) and a fixed concentration of FB (1 μ
m
) flowed over immobilized C3b. Steady-state analysis of the binding data was used to derive dissociation constants for FD binding to C3bB. C, AFD blocked binding of FD to the C3bB complex. Fixed concentrations of FB (1 μ
m
) and FD(S195A) (1 μ
m
) and increasing concentrations of AFD or 8E2 were injected over immobilized C3b. The IC50 value averaged over three separate experiments is 0.43 ± 0.20 μ
m
(mean ± S.D.) with full blockade at 0.82 ± 0.18 μ
m
, which indicates that AFD blocks proconvertase activation at an ∼1:1 molar ratio of AFD to FD. R.U., resonance units.
FIGURE 3.
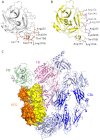
Crystal structure of human FD in complex with AFD. A, the structure of one of the two FD-AFD complexes in the asymmetric unit is shown as a ribbon diagram. FD is shown in white, and the AFD LC and HC are shown in yellow and orange, respectively. VD, variable domain; CD, constant domain. B, close-up of the FD catalytic triad His-57, Asp-102, and Ser-195. The structures of human FD (green) and cynomolgus FD (white) are overlaid. His-57 is shown in three different positions as found in different structures. Ser-215 of the self-inhibitory loop is also indicated.
FIGURE 4.
Interface between AFD and FD. A, FD (transparent surface) interacts with the AFD HC mainly through residues in the 170 loop (orange) and interacts with both the HC and LC through residues in the 220 loop (yellow). B, Arg-170A of FD forms six hydrogen bonds with LC and HC residues of AFD. Potential hydrogen bonds are formed between Lys-223A of FD and Asp-30 and Asp-32 of the AFD LC and between Arg-170A of FD and Tyr-36 and Asn-34 of the AFD LC as well Glu-97 and Glu-99 of the AFD HC.
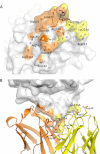
FIGURE 5.
AFD sterically prevents FD from binding to C3bB proconvertase. A, residues in FD (white ribbon representation, from the FD-AFD complex) in contact with AFD (orange sticks). B, residues in FD (yellow ribbon representation, from the C3bBD complex) in contact with FB (pink sticks). C, modeling of a major steric clash between FB (pink) and the AFD HC (orange) and LC (yellow). Dark blue indicates the C3b β-chain, and light blue indicates the C3b α-chain.
Similar articles
- Complement inhibition in cynomolgus monkeys by anti-factor d antigen-binding fragment for the treatment of an advanced form of dry age-related macular degeneration.
Loyet KM, Good J, Davancaze T, Sturgeon L, Wang X, Yang J, Le KN, Wong M, Hass PE, van Lookeren Campagne M, Haughney PC, Morimoto A, Damico-Beyer LA, DeForge LE. Loyet KM, et al. J Pharmacol Exp Ther. 2014 Dec;351(3):527-37. doi: 10.1124/jpet.114.215921. Epub 2014 Sep 17. J Pharmacol Exp Ther. 2014. PMID: 25232192 - Structures of C3b in complex with factors B and D give insight into complement convertase formation.
Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P. Forneris F, et al. Science. 2010 Dec 24;330(6012):1816-20. doi: 10.1126/science.1195821. Science. 2010. PMID: 21205667 Free PMC article. - Structural and functional analysis of a C3b-specific antibody that selectively inhibits the alternative pathway of complement.
Katschke KJ Jr, Stawicki S, Yin J, Steffek M, Xi H, Sturgeon L, Hass PE, Loyet KM, Deforge L, Wu Y, van Lookeren Campagne M, Wiesmann C. Katschke KJ Jr, et al. J Biol Chem. 2009 Apr 17;284(16):10473-9. doi: 10.1074/jbc.M809106200. Epub 2009 Feb 5. J Biol Chem. 2009. PMID: 19196712 Free PMC article. - Structural biology of the alternative pathway convertase.
Xu Y, Narayana SV, Volanakis JE. Xu Y, et al. Immunol Rev. 2001 Apr;180:123-35. doi: 10.1034/j.1600-065x.2001.1800111.x. Immunol Rev. 2001. PMID: 11414354 Review. - Activation of the alternative complement pathway.
Fearon DT. Fearon DT. CRC Crit Rev Immunol. 1979 Nov;1(1):1-32. CRC Crit Rev Immunol. 1979. PMID: 162484 Review. No abstract available.
Cited by
- Lymphocytes as liver damage mirror of HCV related adipogenesis deregulation.
Minutolo A, Conti B, Grelli S, Viscomi C, Labbadia G, Balsano C. Minutolo A, et al. PLoS One. 2014 Mar 21;9(3):e92343. doi: 10.1371/journal.pone.0092343. eCollection 2014. PLoS One. 2014. PMID: 24658135 Free PMC article. - The Challenges and Promise of Complement Therapeutics for Ocular Diseases.
Park DH, Connor KM, Lambris JD. Park DH, et al. Front Immunol. 2019 May 15;10:1007. doi: 10.3389/fimmu.2019.01007. eCollection 2019. Front Immunol. 2019. PMID: 31156618 Free PMC article. Review. - Manipulating the mediator: modulation of the alternative complement pathway C3 convertase in health, disease and therapy.
Ricklin D. Ricklin D. Immunobiology. 2012 Nov;217(11):1057-66. doi: 10.1016/j.imbio.2012.07.016. Immunobiology. 2012. PMID: 22964231 Free PMC article. Review. - Validating therapeutic targets through human genetics.
Plenge RM, Scolnick EM, Altshuler D. Plenge RM, et al. Nat Rev Drug Discov. 2013 Aug;12(8):581-94. doi: 10.1038/nrd4051. Epub 2013 Jul 19. Nat Rev Drug Discov. 2013. PMID: 23868113 Review. - New milestones ahead in complement-targeted therapy.
Ricklin D, Lambris JD. Ricklin D, et al. Semin Immunol. 2016 Jun;28(3):208-22. doi: 10.1016/j.smim.2016.06.001. Epub 2016 Jun 16. Semin Immunol. 2016. PMID: 27321574 Free PMC article. Review.
References
- Hecker L. A., Edwards A. O., Ryu E., Tosakulwong N., Baratz K. H., Brown W. L., Charbel Issa P., Scholl H. P., Pollok-Kopp B., Schmid-Kubista K. E., Bailey K. R., Oppermann M. (2010) Genetic control of the alternative pathway of complement in humans and age-related macular degeneration. Hum. Mol. Genet. 19, 209–215 - PMC - PubMed
- Stanton C. M., Yates J. R., den Hollander A. I., Seddon J. M., Swaroop A., Stambolian D., Fauser S., Hoyng C., Yu Y., Atsuhiro K., Branham K., Othman M., Chen W., Kortvely E., Chalmers K., Hayward C., Moore A. T., Dhillon B., Ueffing M., Wright A. F. (2011) Complement factor D in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 52, 8828–8834 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases