Patterns of gene flow define species of thermophilic Archaea - PubMed (original) (raw)
Patterns of gene flow define species of thermophilic Archaea
Hinsby Cadillo-Quiroz et al. PLoS Biol. 2012 Feb.
Abstract
Despite a growing appreciation of their vast diversity in nature, mechanisms of speciation are poorly understood in Bacteria and Archaea. Here we use high-throughput genome sequencing to identify ongoing speciation in the thermoacidophilic Archaeon Sulfolobus islandicus. Patterns of homologous gene flow among genomes of 12 strains from a single hot spring in Kamchatka, Russia, demonstrate higher levels of gene flow within than between two persistent, coexisting groups, demonstrating that these microorganisms fit the biological species concept. Furthermore, rates of gene flow between two species are decreasing over time in a manner consistent with incipient speciation. Unlike other microorganisms investigated, we do not observe a relationship between genetic divergence and frequency of recombination along a chromosome, or other physical mechanisms that would reduce gene flow between lineages. Each species has its own genetic island encoding unique physiological functions and a unique growth phenotype that may be indicative of ecological specialization. Genetic differentiation between these coexisting groups occurs in large genomic "continents," indicating the topology of genomic divergence during speciation is not uniform and is not associated with a single locus under strong diversifying selection. These data support a model where species do not require physical barriers to gene flow but are maintained by ecological differentiation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
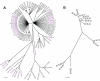
Figure 1. Phylogenetic relationships among S. islandicus from the Mutnovsky Volcano.
(A) ClonalFrame reconstruction based on seven loci from 97 S. islandicus strains from the Mutnovsky Volcano region of Kamchatka, Russia (details in Table S1). Strains in purple were isolated from spring M.16. The first number in each name indicates the spring from which strains were isolated, the second indicates the isolate number from that spring, and the third indicates year of isolation. * designates strains selected for genome sequencing and comparison. (B) ClonalFrame phylogeny based on the core genome alignment of 12 S. islandicus strains from hot spring M.16.
Figure 2. Heat map representation of homologous recombination frequency for every donor/recipient pair of branches of the core genome phylogeny of 12 S. islandicus strains.
Recombination frequency is measured relative to its expectation under the prior of the ClonalOrigin model and color coded according to the upper left color/magnitude legend (light blue and blue for the frequency of recombination events below a 1∶1 ratio and yellow to red for the frequency of recombination events above 1∶1). Light gray cells represent non-significant ratios with less than four observed and expected events. White shows number of events that match the prior expectations. Names of strains are color coded as Blue and Red groups.
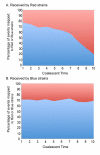
Figure 3. Variation in recombination events between the Red and Blue groups through time.
For the Red (A) and Blue (B) recipient strains, the total proportion of events that could be assigned as originating from either donor Red (colored red) and Blue (colored blue) strains is shown as a function of coalescent time with 10 being the most recent divergence and 1 being the common ancestor of this set of strains. A coalescent unit of time is equal to the average length of a generation multiplied by the effective population size.
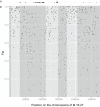
Figure 4. FST values between the Red and Blue groups along the chromosome of strain M.16.27.
(A) 10,000 bp windows on the M.16.27 genome where genome sequence is not present in all 10 strains from the Red and Blue groups. These positions highlight variable portions of the M.16.27 genome. *Indicates a recently integrated plasmid. (B) FST values were calculated for sliding windows of 10 kb moving in 5 kb steps. Empty windows where sequence from M.16.27 is not shared by all strains are not plotted. Shading highlights regions of the chromosome that are less differentiated beginning and ending with the first window with FST values lower than 0.5.
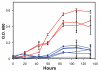
Figure 5. Growth of M.16 strains under standard heterotrophic conditions.
Lines are color coded for strains assigned to the Red and Blue groups. Negative control with no inoculum added is shown in grey. Error bars show the variation in growth among three independent replicate cultures.
Similar articles
- Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus.
Whitaker RJ, Grogan DW, Taylor JW. Whitaker RJ, et al. Mol Biol Evol. 2005 Dec;22(12):2354-61. doi: 10.1093/molbev/msi233. Epub 2005 Aug 10. Mol Biol Evol. 2005. PMID: 16093568 - Adaptive divergence with gene flow in incipient speciation of Miscanthus floridulus/sinensis complex (Poaceae).
Huang CL, Ho CW, Chiang YC, Shigemoto Y, Hsu TW, Hwang CC, Ge XJ, Chen C, Wu TH, Chou CH, Huang HJ, Gojobori T, Osada N, Chiang TY. Huang CL, et al. Plant J. 2014 Dec;80(5):834-47. doi: 10.1111/tpj.12676. Epub 2014 Nov 11. Plant J. 2014. PMID: 25237766 - Biogeography of the Sulfolobus islandicus pan-genome.
Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. Reno ML, et al. Proc Natl Acad Sci U S A. 2009 May 26;106(21):8605-10. doi: 10.1073/pnas.0808945106. Epub 2009 May 12. Proc Natl Acad Sci U S A. 2009. PMID: 19435847 Free PMC article. - Sulfolobus islandicus: a model system for evolutionary genomics.
Zhang C, Krause DJ, Whitaker RJ. Zhang C, et al. Biochem Soc Trans. 2013 Feb 1;41(1):458-62. doi: 10.1042/BST20120338. Biochem Soc Trans. 2013. PMID: 23356328 Review. - Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow.
Cruickshank TE, Hahn MW. Cruickshank TE, et al. Mol Ecol. 2014 Jul;23(13):3133-57. doi: 10.1111/mec.12796. Epub 2014 Jun 17. Mol Ecol. 2014. PMID: 24845075 Review.
Cited by
- A Latitudinal Diversity Gradient in Terrestrial Bacteria of the Genus Streptomyces.
Andam CP, Doroghazi JR, Campbell AN, Kelly PJ, Choudoir MJ, Buckley DH. Andam CP, et al. mBio. 2016 Apr 12;7(2):e02200-15. doi: 10.1128/mBio.02200-15. mBio. 2016. PMID: 27073097 Free PMC article. - Horizontal gene transfer, dispersal and haloarchaeal speciation.
Papke RT, Corral P, Ram-Mohan N, de la Haba RR, Sánchez-Porro C, Makkay A, Ventosa A. Papke RT, et al. Life (Basel). 2015 May 19;5(2):1405-26. doi: 10.3390/life5021405. Life (Basel). 2015. PMID: 25997110 Free PMC article. Review. - The Genome of a Thermo Tolerant, Pathogenic Albino Aspergillus fumigatus.
Couger B, Weirick T, Damásio ARL, Segato F, Polizeli MLTM, de Almeida RSC, Goldman GH, Prade RA. Couger B, et al. Front Microbiol. 2018 Aug 14;9:1827. doi: 10.3389/fmicb.2018.01827. eCollection 2018. Front Microbiol. 2018. PMID: 30154766 Free PMC article. - Diversified local CRISPR-Cas immunity to viruses of Sulfolobus islandicus.
Pauly MD, Bautista MA, Black JA, Whitaker RJ. Pauly MD, et al. Philos Trans R Soc Lond B Biol Sci. 2019 May 13;374(1772):20180093. doi: 10.1098/rstb.2018.0093. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 30905292 Free PMC article. - Genomic and phenotypic differentiation among Methanosarcina mazei populations from Columbia River sediment.
Youngblut ND, Wirth JS, Henriksen JR, Smith M, Simon H, Metcalf WW, Whitaker RJ. Youngblut ND, et al. ISME J. 2015 Oct;9(10):2191-205. doi: 10.1038/ismej.2015.31. Epub 2015 Mar 10. ISME J. 2015. PMID: 25756680 Free PMC article.
References
- Acinas S. G, Klepac-Ceraj V, Hunt D. E, Pharino C, Ceraj I, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. - PubMed
- Hunt D. E, David L. A, Gevers D, Preheim S. P, Alm E. J, et al. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320(5879):1081–1085. - PubMed
- Oakley B. B, Carbonero F, van der Gast C. J, Hawkins R. J, Purdy K. J. Evolutionary divergence and biogeography of sympatric niche-differentiated bacterial populations. ISME Journal. 2010;4:488–497. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases