Identification of gene-oriented exon orthology between human and mouse - PubMed (original) (raw)
Identification of gene-oriented exon orthology between human and mouse
Gloria C-L Fu et al. BMC Genomics. 2012.
Abstract
Background: Gene orthology has been well studied in the evolutionary area and is thought to be an important implication to functional genome annotations. As the accumulation of transcriptomic data, alternative splicing is taken into account in the assignments of gene orthologs and the orthology is suggested to be further considered at transcript level. Whether gene or transcript orthology, exons are the basic units that represent the whole gene structure; however, there is no any reported study on how to build exon level orthology in a whole genome scale. Therefore, it is essential to establish a gene-oriented exon orthology dataset.
Results: Using a customized pipeline, we first build exon orthologous relationships from assigned gene orthologs pairs in two well-annotated genomes: human and mouse. More than 92% of non-overlapping exons have at least one ortholog between human and mouse and only a small portion of them own more than one ortholog. The exons located in the coding region are more conserved in terms of finding their ortholog counterparts. Within the untranslated region, the 5' UTR seems to have more diversity than the 3' UTR according to exon orthology designations. Interestingly, most exons located in the coding region are also conserved in length but this conservation phenomenon dramatically drops down in untranslated regions. In addition, we allowed multiple assignments in exon orthologs and a subset of exons with possible fusion/split events were defined here after a thorough analysis procedure.
Conclusions: Identification of orthologs at the exon level is essential to provide a detailed way to interrogate gene orthology and splicing analysis. It could be used to extend the genome annotation as well. Besides examining the one-to-one orthologous relationship, we manage the one-to-multi exon pairs to represent complicated exon generation behavior. Our results can be further applied in many research fields studying intron-exon structure and alternative/constitutive exons in functional genomic areas.
Figures
Figure 1
Process of generating human and mouse exon orthologs. A. Flow chart of orthologous exon database building. B. Exons with N (N > 1) putative exon orthologs were further verified if these N exons belonged to the same united exon. C. Identification of fused and split exons through analysis procedure from putative orthologous exons. D. Confirmation of the correct orthologous exon pairs via anchor mapping for the rest of the putative orthologs yet to be verified.
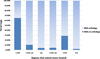
Figure 2
Distribution of united exons with and without orthologous pairs in different gene regions. The x-axis represents the gene regions in which united exons are located, and the y-axis is the percentage of united exons with or without orthologs to the total number of united exons in each category. SLR means a single long united exon extending from 5' UTR to 3' UTR.
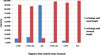
Figure 3
Comparisons of united exons with or without length conservation to their orthologous pairs from human and mouse. Only one-to-one orthologous pairs are analyzed in this figure. The x-axis indicates the gene regions in which the united exons are located. The SLR tag means that the united exon crossed through 5' UTR, the coding region, and 3' UTR. The y-axis represents the number of united exons in two categories (with equal or unequal length to orthologous pairs) normalized to the total number of united exons in each gene region.
Figure 4
Analysis of length differences between orthologous pairs of unequal exon length. A. Distribution of remainders of three in length differences between orthologous exon pairs with unequal length. The MOD 3 = 0 category means that the remainder is zero when the length difference between exon orthologs is divided by three, and so on. The x-axis is the region in which the united exon is located, and the y-axis indicates the percentage of orthologous pairs with unequal length in each region for each category. B. Distribution in counts of united exons where length difference is less than or equal to 30 base pairs compared to orthologous exon pairs.
Similar articles
- SplicedFamAlign: CDS-to-gene spliced alignment and identification of transcript orthology groups.
Jammali S, Aguilar JD, Kuitche E, Ouangraoua A. Jammali S, et al. BMC Bioinformatics. 2019 Mar 29;20(Suppl 3):133. doi: 10.1186/s12859-019-2647-2. BMC Bioinformatics. 2019. PMID: 30925859 Free PMC article. - Identifying genes with conserved splicing structure and orthologous isoforms in human, mouse and dog.
Guillaudeux N, Belleannée C, Blanquart S. Guillaudeux N, et al. BMC Genomics. 2022 Mar 18;23(1):216. doi: 10.1186/s12864-022-08429-4. BMC Genomics. 2022. PMID: 35303798 Free PMC article. - Assessment of orthologous splicing isoforms in human and mouse orthologous genes.
Zambelli F, Pavesi G, Gissi C, Horner DS, Pesole G. Zambelli F, et al. BMC Genomics. 2010 Oct 1;11:534. doi: 10.1186/1471-2164-11-534. BMC Genomics. 2010. PMID: 20920313 Free PMC article. - The Protein-Coding Human Genome: Annotating High-Hanging Fruits.
Hatje K, Mühlhausen S, Simm D, Kollmar M. Hatje K, et al. Bioessays. 2019 Nov;41(11):e1900066. doi: 10.1002/bies.201900066. Epub 2019 Sep 23. Bioessays. 2019. PMID: 31544971 Review. - Advances in the Exon-Intron Database (EID).
Shepelev V, Fedorov A. Shepelev V, et al. Brief Bioinform. 2006 Jun;7(2):178-85. doi: 10.1093/bib/bbl003. Epub 2006 Mar 9. Brief Bioinform. 2006. PMID: 16772261 Review.
Cited by
- Detection of orthologous exons and isoforms using EGIO.
Ma J, Wu JY, Zhu L. Ma J, et al. Bioinformatics. 2022 Sep 30;38(19):4474-4480. doi: 10.1093/bioinformatics/btac548. Bioinformatics. 2022. PMID: 35946527 Free PMC article. - The Evolution of Hemocyanin Genes in Caenogastropoda: Gene Duplications and Intron Accumulation in Highly Diverse Gastropods.
Schäfer GG, Grebe LJ, Schinkel R, Lieb B. Schäfer GG, et al. J Mol Evol. 2021 Dec;89(9-10):639-655. doi: 10.1007/s00239-021-10036-y. Epub 2021 Nov 10. J Mol Evol. 2021. PMID: 34757470 Free PMC article. - Functional divergence and intron variability during evolution of angiosperm TERMINAL FLOWER1 (TFL1) genes.
Gao J, Huang BH, Wan YT, Chang J, Li JQ, Liao PC. Gao J, et al. Sci Rep. 2017 Nov 1;7(1):14830. doi: 10.1038/s41598-017-13645-0. Sci Rep. 2017. PMID: 29093470 Free PMC article. - RNA-Seq Differentiates Tumour and Host mRNA Expression Changes Induced by Treatment of Human Tumour Xenografts with the VEGFR Tyrosine Kinase Inhibitor Cediranib.
Bradford JR, Farren M, Powell SJ, Runswick S, Weston SL, Brown H, Delpuech O, Wappett M, Smith NR, Carr TH, Dry JR, Gibson NJ, Barry ST. Bradford JR, et al. PLoS One. 2013 Jun 19;8(6):e66003. doi: 10.1371/journal.pone.0066003. Print 2013. PLoS One. 2013. PMID: 23840389 Free PMC article.
References
- Bafna V, Huson DH. The conserved exon method for gene finding. Proc Int Conf Intell Syst Mol Biol. 2000;8:3–12. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials