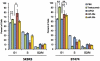Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells - PubMed (original) (raw)
Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells
Takehiro Ichikawa et al. PLoS One. 2012.
Abstract
Objective: Trastuzumab has been used for the treatment of HER2-positive breast cancer (BC). However, a subset of BC patients exhibited resistance to trastuzumab therapy. Thus, clarifying the molecular mechanism of trastuzumab treatment will be beneficial to improve the treatment of HER2-positive BC patients. In this study, we identified trastuzumab-responsive microRNAs that are involved in the therapeutic effects of trastuzumab.
Methods and results: RNA samples were obtained from HER2-positive (SKBR3 and BT474) and HER2-negetive (MCF7 and MDA-MB-231) cells with and without trastuzumab treatment for 6 days. Next, we conducted a microRNA profiling analysis using these samples to screen those microRNAs that were up- or down-regulated only in HER2-positive cells. This analysis identified miR-26a and miR-30b as trastuzumab-inducible microRNAs. Transfecting miR-26a and miR-30b induced cell growth suppression in the BC cells by 40% and 32%, respectively. A cell cycle analysis showed that these microRNAs induced G1 arrest in HER2-positive BC cells as trastuzumab did. An Annexin-V assay revealed that miR-26a but not miR-30b induced apoptosis in HER2-positive BC cells. Using the prediction algorithms for microRNA targets, we identified cyclin E2 (CCNE2) as a target gene of miR-30b. A luciferase-based reporter assay demonstrated that miR-30b post-transcriptionally reduced 27% (p = 0.005) of the gene expression by interacting with two binding sites in the 3'-UTR of CCNE2.
Conclusion: In BC cells, trastuzumab modulated the expression of a subset of microRNAs, including miR-26a and miR-30b. The upregulation of miR-30b by trastuzumab may play a biological role in trastuzumab-induced cell growth inhibition by targeting CCNE2.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
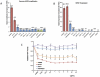
Figure 1. HER2-status of breast cancer cell lines.
The genomic amplification (1A) and mRNA expression level (1B) of HER2 in 11 human breast cancer cell lines and normal human mammary epithelial cells (HMEC) were assessed using quantitative PCR and quantitative RT-PCR (n = 3). The mRNA abundance was normalized by the GAPDH expression levels. 1C: The trastuzumab sensitivity of SKBR3, BT474, MCF7, and MDA-MB-231 cells was determined using the WST-1 assay. The cells were incubated in trastuzumab-containing media at different concentrations for 144 hours, and then the absorbance at 450 nm was measured after a 2-hour incubation with WST-1 reagent. The ratio of the absorbance to that of the non-treated cells represented the trastuzumab sensitivity of cells.
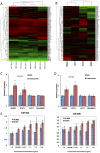
Figure 2. Identification of trastuzumab-responsive microRNAs.
2A: A heat map and clustergram of the expression profile of 71 pre-filtered microRNAs. The red and green represent higher and lower expression levels, respectively. (tras +): with trastuzumab treatment, (tras −): without trastuzumab treatment. 2B: A heatmap and clustergram of the fold-change of microRNA expression by trastuzumab treatment. The red and green represent up- and down-regulation. 2C and 2D: The expression levels of miR-26a (2C) and miR-30b (2D) were validated by qRT-PCR (n = 3). The data are shown as microRNA expression levels relative to a control treatment (PBS). 2E: The expression level of miR-26a and miR-30b in different trastuzumab concentrations was measured (n = 2). The microRNA expression levels were normalized against miR-16. All bars and error bars represent means ± SEM. *: p<0.05.
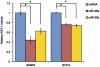
Figure 3. Effects of miR-26a and miR-30b on cell proliferation.
The cells were transfected with negative control RNA (ncRNA), miR-26a, or miR-30b. At 72 hours after the transfection, the amount of viable cells was assessed by the WST-1 assay. The WST-1 activity values were normalized against that of the ncRNA-treatment. All bars and error bars represent means ± SEM (n = 4). *: p<0.05.
Figure 4. Effects of miR-26a and miR-30b on the cell cycle.
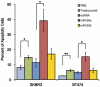
The PI-stained DNA content of the cells was evaluated using a FACS Calibur (BD Biosciences) at 72 hours after transfection. All bars and error bars represent means ± SEM (n = 6). *: p<0.05, **: p<0.005.
Figure 5. Effects of miR-26a and miR-30b on apoptosis.
The apoptotic cells were detected using FITC-Annexin V at 72 hours after microRNA transfection. The percentage of Annexin V-FITC positive cells to the total cells was shown in the bar graphs. All bars and error bars represent means ± SEM (n = 4). *: p<0.05, **: p<0.005.
Figure 6. CCNE2 is a direct target of miR-30b in breast cancer cells.
6A: A diagram of the 3′UTR-containing reporter constructs for CCNE2, CCNA1, and CDC7 and their derivatives. The 3′UTRs of the three genes were inserted just downstream of the firefly luciferase gene in the pGL4.13 vector (wt). Next, the mutated derivatives (mut1, mut2, and mut1+2) of CCNE2-wt were generated by inserting mutations into two putative binding sites corresponding to the seed-sequence of miR-30b. 6B and 6C: SKBR3 and BT474 cells were co-transfected with reporter constructs, internal control vector (pGL4.73), and synthetic miR-30b oligomer. 6D: assessment of endogenous microRNA's inhibitory effects to CCNE2. Only reporter constructs and pGL4.73 were transfected into SKBR3 and BT474 cells. Twenty-four hours after the transfection, the reporter luciferase activity was measured. The data were shown as the luciferase activity relative to that of vehicle (pGL4.13+pGL4.73) transfection. All bars and error bars represent means ± SEM (n = 3). *: p<0.05, **: p<0.005.
Similar articles
- The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene.
Tormo E, Adam-Artigues A, Ballester S, Pineda B, Zazo S, González-Alonso P, Albanell J, Rovira A, Rojo F, Lluch A, Eroles P. Tormo E, et al. Sci Rep. 2017 Jan 25;7:41309. doi: 10.1038/srep41309. Sci Rep. 2017. PMID: 28120942 Free PMC article. - Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer.
Le XF, Almeida MI, Mao W, Spizzo R, Rossi S, Nicoloso MS, Zhang S, Wu Y, Calin GA, Bast RC Jr. Le XF, et al. PLoS One. 2012;7(7):e41170. doi: 10.1371/journal.pone.0041170. Epub 2012 Jul 19. PLoS One. 2012. PMID: 22829924 Free PMC article. - Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer.
Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, Su F, Yao H, Song E. Gong C, et al. J Biol Chem. 2011 May 27;286(21):19127-37. doi: 10.1074/jbc.M110.216887. Epub 2011 Apr 6. J Biol Chem. 2011. PMID: 21471222 Free PMC article. - HER2-targeting antibodies modulate the cyclin-dependent kinase inhibitor p27Kip1 via multiple signaling pathways.
Le XF, Pruefer F, Bast RC Jr. Le XF, et al. Cell Cycle. 2005 Jan;4(1):87-95. doi: 10.4161/cc.4.1.1360. Epub 2005 Jan 10. Cell Cycle. 2005. PMID: 15611642 Review. - MicroRNA and HER2-overexpressing cancer.
Wang SE, Lin RJ. Wang SE, et al. Microrna. 2013;2(2):137-47. doi: 10.2174/22115366113029990011. Microrna. 2013. PMID: 25070783 Free PMC article. Review.
Cited by
- 3, 3'-Diindolylmethane enhances the effectiveness of herceptin against HER-2/neu-expressing breast cancer cells.
Ahmad A, Ali S, Ahmed A, Ali AS, Raz A, Sakr WA, Rahman KM. Ahmad A, et al. PLoS One. 2013;8(1):e54657. doi: 10.1371/journal.pone.0054657. Epub 2013 Jan 22. PLoS One. 2013. PMID: 23372748 Free PMC article. - Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing.
Aganezov S, Goodwin S, Sherman RM, Sedlazeck FJ, Arun G, Bhatia S, Lee I, Kirsche M, Wappel R, Kramer M, Kostroff K, Spector DL, Timp W, McCombie WR, Schatz MC. Aganezov S, et al. Genome Res. 2020 Sep;30(9):1258-1273. doi: 10.1101/gr.260497.119. Epub 2020 Sep 4. Genome Res. 2020. PMID: 32887686 Free PMC article. - miRNA-26a blocks interleukin-2-mediated migration and proliferation of non-small cell lung cancer cells via vascular cell adhesion molecule-1.
Li L, Li D, Chen Y. Li L, et al. Transl Cancer Res. 2020 Mar;9(3):1768-1778. doi: 10.21037/tcr.2020.02.36. Transl Cancer Res. 2020. PMID: 35117524 Free PMC article. - Involvement of miR-30b in kynurenine-mediated lysyl oxidase expression.
Duan Z, Li L, Li Y. Duan Z, et al. J Physiol Biochem. 2019 Jun;75(2):135-142. doi: 10.1007/s13105-019-00686-4. Epub 2019 May 15. J Physiol Biochem. 2019. PMID: 31093946 - ER Negative Breast Cancer and miRNA: There Is More to Decipher Than What the Pathologist Can See!
Chamandi G, El-Hajjar L, El Kurdi A, Le Bras M, Nasr R, Lehmann-Che J. Chamandi G, et al. Biomedicines. 2023 Aug 18;11(8):2300. doi: 10.3390/biomedicines11082300. Biomedicines. 2023. PMID: 37626796 Free PMC article. Review.
References
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. - PubMed
- Hynes NE, Stern DF. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim Biophys Acta. 1994;1198:165–184. - PubMed
- Rosen PP, Lesser ML, Arroyo CD, Cranor M, Borgen P, et al. Immunohistochemical detection of HER2/neu in patients with axillary lymph node negative breast carcinoma. A study of epidemiologic risk factors, histologic features, and prognosis. Cancer. 1995;75:1320–1326. - PubMed
- Carlomagno C, Perrone F, Gallo C, De Laurentiis M, Lauria R, et al. c-erb B2 overexpression decreases the benefit of adjuvant tamoxifen in early-stage breast cancer without axillary lymph node metastases. J Clin Oncol. 1996;14:2702–2708. - PubMed
- Gusterson BA, Gelber RD, Goldhirsch A, Price KN, Save-Soderborgh J, et al. Prognostic importance of c-erbB-2 expression in breast cancer. International (Ludwig) Breast Cancer Study Group. J Clin Oncol. 1992;10:1049–1056. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous