Fast gapped-read alignment with Bowtie 2 - PubMed (original) (raw)
Fast gapped-read alignment with Bowtie 2
Ben Langmead et al. Nat Methods. 2012.
Abstract
As the rate of sequencing increases, greater throughput is demanded from read aligners. The full-text minute index is often used to make alignment very fast and memory-efficient, but the approach is ill-suited to finding longer, gapped alignments. Bowtie 2 combines the strengths of the full-text minute index with the flexibility and speed of hardware-accelerated dynamic programming algorithms to achieve a combination of high speed, sensitivity and accuracy.
Figures
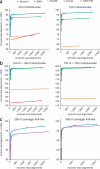
Figure 1
Alignment comparison using HiSeq 2000, 454 and Ion Torrent reads. (a–d) Bowtie 2, BWA, SOAP2 and Bowtie were used to align two million 100 nt × 100 nt paired-end HiSeq 2000 reads from a resequencing study. Shown are results for unpaired alignment of end 1 (a), paired-end alignment (b), Bowtie 2 and BWA-SW alignment of 1 million 454 reads from the 1000 Genomes Project Pilot (c), and Bowtie 2 and BWA-SW to align one million Ion Torrent reads from the G. Moore resequencing project (d). Plotted is the percentage of reads for which at least one alignment was found. Each numbered point is data obtained using command-line parameters shown in Supplementary Table 1.
Figure 2
Sensitivity and accuracy of alignment using simulated reads. (a–c) Cumulative number of correct and incorrect alignments from high to low mapping quality for simulated Illumina-like unpaired 100 nt and 150 nt datasets (a), for simulated Illumina-like paired-end 100 nt × 100 nt and 150 nt × 150 nt datasets (b), and for simulated 454-like datasets with average read lengths 250 nt and 400 nt (c) using indicated aligners.
Similar articles
- The GEM mapper: fast, accurate and versatile alignment by filtration.
Marco-Sola S, Sammeth M, Guigó R, Ribeca P. Marco-Sola S, et al. Nat Methods. 2012 Dec;9(12):1185-8. doi: 10.1038/nmeth.2221. Epub 2012 Oct 28. Nat Methods. 2012. PMID: 23103880 - CUSHAW: a CUDA compatible short read aligner to large genomes based on the Burrows-Wheeler transform.
Liu Y, Schmidt B, Maskell DL. Liu Y, et al. Bioinformatics. 2012 Jul 15;28(14):1830-7. doi: 10.1093/bioinformatics/bts276. Epub 2012 May 9. Bioinformatics. 2012. PMID: 22576173 - X-Mapper: fast and accurate sequence alignment via gapped x-mers.
Gaston JM, Alm EJ, Zhang AN. Gaston JM, et al. Genome Biol. 2025 Jan 22;26(1):15. doi: 10.1186/s13059-024-03473-7. Genome Biol. 2025. PMID: 39844205 Free PMC article. - Technology dictates algorithms: recent developments in read alignment.
Alser M, Rotman J, Deshpande D, Taraszka K, Shi H, Baykal PI, Yang HT, Xue V, Knyazev S, Singer BD, Balliu B, Koslicki D, Skums P, Zelikovsky A, Alkan C, Mutlu O, Mangul S. Alser M, et al. Genome Biol. 2021 Aug 26;22(1):249. doi: 10.1186/s13059-021-02443-7. Genome Biol. 2021. PMID: 34446078 Free PMC article. Review. - Performance optimization in DNA short-read alignment.
Wilton R, Szalay AS. Wilton R, et al. Bioinformatics. 2022 Apr 12;38(8):2081-2087. doi: 10.1093/bioinformatics/btac066. Bioinformatics. 2022. PMID: 35139149 Free PMC article. Review.
Cited by
- GWAS for Drought Resilience Traits in Red Clover (Trifolium pratense L.).
Vleugels T, Ruttink T, Ariza-Suarez D, Dubey R, Saleem A, Roldán-Ruiz I, Muylle H. Vleugels T, et al. Genes (Basel). 2024 Oct 21;15(10):1347. doi: 10.3390/genes15101347. Genes (Basel). 2024. PMID: 39457472 Free PMC article. - Investigating Anthrax-Associated Virulence Genes among Archival and Contemporary Bacillus cereus Group Genomes.
Sabin SJ, Beesley CA, Marston CK, Paisie TK, Gulvik CA, Sprenger GA, Gee JE, Traxler RM, Bell ME, McQuiston JR, Weiner ZP. Sabin SJ, et al. Pathogens. 2024 Oct 10;13(10):884. doi: 10.3390/pathogens13100884. Pathogens. 2024. PMID: 39452755 Free PMC article. - Novel pharmacologic inhibition of lysine-specific demethylase 1 as a potential therapeutic for glioblastoma.
Shinjo K, Umehara T, Niwa H, Sato S, Katsushima K, Sato S, Wang X, Murofushi Y, Suzuki MM, Koyama H, Kondo Y. Shinjo K, et al. Cancer Gene Ther. 2024 Dec;31(12):1884-1894. doi: 10.1038/s41417-024-00847-8. Epub 2024 Nov 5. Cancer Gene Ther. 2024. PMID: 39501082 Free PMC article. - Oral Microbial Translocation Genes in Gastrointestinal Cancers: Insights from Metagenomic Analysis.
Wang L, Wang Q, Zhou Y. Wang L, et al. Microorganisms. 2024 Oct 18;12(10):2086. doi: 10.3390/microorganisms12102086. Microorganisms. 2024. PMID: 39458395 Free PMC article. - Construction of a High-Density Genetic Map of Acca sellowiana (Berg.) Burret, an Outcrossing Species, Based on Two Connected Mapping Populations.
Quezada M, Amadeu RR, Vignale B, Cabrera D, Pritsch C, Garcia AAF. Quezada M, et al. Front Plant Sci. 2021 Feb 23;12:626811. doi: 10.3389/fpls.2021.626811. eCollection 2021. Front Plant Sci. 2021. PMID: 33708232 Free PMC article.
References
- Ferragina P, Manzini G. Proc. 41st Annual Symposium on Foundations of Computer Science. IEEE Comput. Soc.; 2000. pp. 390–398.
Publication types
MeSH terms
Grants and funding
- R01 HG006677/HG/NHGRI NIH HHS/United States
- R01-HG006102/HG/NHGRI NIH HHS/United States
- R01 HG006102-02/HG/NHGRI NIH HHS/United States
- R01 HG006677-12/HG/NHGRI NIH HHS/United States
- R01 HG006102/HG/NHGRI NIH HHS/United States
- R01 HG006677-13/HG/NHGRI NIH HHS/United States
- R01-GM083873/GM/NIGMS NIH HHS/United States
- R01 GM083873/GM/NIGMS NIH HHS/United States
- R01 HG006102-01/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials