Regulation of cytoplasmic mRNA decay - PubMed (original) (raw)
Review
Regulation of cytoplasmic mRNA decay
Daniel R Schoenberg et al. Nat Rev Genet. 2012.
Erratum in
- Nat Rev Genet. 2012 Jun;13(6):448
Abstract
Discoveries made over the past 20 years highlight the importance of mRNA decay as a means of modulating gene expression and thereby protein production. Up until recently, studies largely focused on identifying cis-acting sequences that serve as mRNA stability or instability elements, the proteins that bind these elements, how the process of translation influences mRNA decay and the ribonucleases that catalyse decay. Now, current studies have begun to elucidate how the decay process is regulated. This Review examines our current understanding of how mammalian cell mRNA decay is controlled by different signalling pathways and lays out a framework for future research.
Figures
Box 1
Box 4
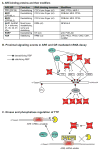
Figure 1. NMD factors as regulators of NMD
NMD in mammalian cells occurs during the translation of mRNA that is bound at its 5′ m7Gppp cap by CBP80 and CBP20. STEP1 involves a weak and/or transient interaction of UPF1 with cap-bound CBP80 – an interaction that typifies all newly synthesized mRNAs. In STEP2, the suppressor of morphogenetic effect on genitalia (SMG)1, which is a PIK-related serine-threonine protein kinase that phosphorylates multiple residues in the C-terminal region of UPF1, binds together with UPF1 to the two translation termination factors, eukaryotic release factor (eRF)1 and eRF3, to form SURF at a PTC. During the formation of the SURF complex SMG1 kinase activity is suppressed by SMG8 via the so-called SMG1 complex (SMG1C). In STEP3 SMG1-UPF1 is joined to an EJC that resides downstream of the PTC. This is dependent upon UPF3X out-competing its functionally less-effective paralog UPF3 for binding at the exon-exon junction complex. The SMG1-mediated phosphorylation of UPF1 that occurs in STEP 4 occurs as a consequence of SMG1-UPF1 binding directly to the UPF2 EJC constituent. Phosphorylated UPF1 binds to eIF3, should a pre-initiation complex form at the AUG translation initiation codon inhibiting additional rounds of translation initiation. In STEP5, SMG6 binds UPF1 dependent on the phosphorylation of UPF1 at threonine 28, and SMG5 and SMG7 bind UPF1 dependent on UPF1 phosphorylation at serine 1096. In STEP 6A, the SMG5- and SMG7-mediated decay of the NMD target occurs by decapping and/or deadenylation followed in STEP 7A by degradation of the body of the transcript in the 5′-to-3′ direction from the decapped 5′-end and/or 3′-to-5′direction from the 3′-deadenylated end. A parallel pathway in STEP 6B involves the SMG6-mediated endonucleolytic cleavage of the NMD target and the accompanying rapid decay of the resulting 5′-cleavage product and 3′-cleavage product in STEP 7B. Decay of the 3′-cleavage product requires mRNP disassembly that is mediated by UPF1.
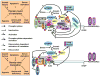
Figure 2. Signaling pathways that regulate the pioneer round of translation and NMD
The pioneer-round of translation utilizes newly synthesized mRNA bound by the cap-binding protein heterodimer CBP80-CBP20 (CBC) and, provided the mRNA derived from splicing, at least one exon-junction complex (EJC, which consists of but is not limited to MAGOH, Y14, PYM and eIF4AIII) situated ~ 20–24-nucleotides upstream of such a junction. This round of translation (and, thus, NMD for those mRNAs that are NMD targets) is promoted upon activation of the mTORC signaling pathway, i.e, in the absence of stress and the presence of growth factors, by multiple pathways. mTOR-mediated translational activation is best understood for eIF4E-bound mRNAs. Translational activation, and thus the activation of any associated mRNA decay pathways, involves mTORC1 binding to eIF3, which results in the phosphorylation of the 4E-BP1 translational repressor and consequently the dissociation of inactivated 4E-BP1 from mRNA-bound eIF4E. eIF4G can then interact with and stabilize the binding of eIF4E, eIF4A and PABPC1 to mRNA to ensure efficient translation. mTORC1 binding to eIF3 also results in the phosphorylation of the S6 kinase 1 (S6K1) translational activator and consequently the dissociation of activated S6K1 from mRNA-bound eIF3. Dissociated S6K1 phosphorylates eIF4B and tumor suppressor programmed cell death 4 (PDCD4), which augment scanning of the 43S preinitiation complex to the translation initiation codon. Taking cues from how mTOR activates eIF4E-bound mRNA translation, the mTOR-mediated translational activation of CBC-bound mRNA, and thus the activation of NMD, may also involve mTORC1 binding to eIF3 and the dissociation of activated S6K1 from CBC-bound mRNA. Activated S6K1 is then recruited by the EJC constituent SKAR, which promotes the pioneer round of translation by enhancing the phosphorylation of SKAR and other downstream effectors that may include eIF4B and PDCD4, which associate with not only eIF4E-bound mRNA but also CBC-bound mRNA. Occurring in parallel, S6K1 phosphorylation by the Cdc42 kinase may augment binding of CBC to mRNA caps, the splicing factor SFRS1 may initiate S6K1 signaling via mTORC1, and the interaction of PYM with the EJC-core proteins Y14 and MAGOH and the 40S ribosomal subunit all appear to promote the pioneer round of translation and, when appropriate, NMD. (After Figure 2 in.)
Similar articles
- The eukaryotic transcriptional machinery regulates mRNA translation and decay in the cytoplasm.
Dahan N, Choder M. Dahan N, et al. Biochim Biophys Acta. 2013 Jan;1829(1):169-73. doi: 10.1016/j.bbagrm.2012.08.004. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982191 Review. - Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway.
Chen CY, Shyu AB. Chen CY, et al. Mol Cell Biol. 2003 Jul;23(14):4805-13. doi: 10.1128/MCB.23.14.4805-4813.2003. Mol Cell Biol. 2003. PMID: 12832468 Free PMC article. - Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay.
Sato H, Hosoda N, Maquat LE. Sato H, et al. Mol Cell. 2008 Feb 1;29(2):255-62. doi: 10.1016/j.molcel.2007.12.009. Mol Cell. 2008. PMID: 18243119 - Cytoplasmic mRNA turnover and ageing.
Borbolis F, Syntichaki P. Borbolis F, et al. Mech Ageing Dev. 2015 Dec;152:32-42. doi: 10.1016/j.mad.2015.09.006. Epub 2015 Oct 1. Mech Ageing Dev. 2015. PMID: 26432921 Free PMC article. Review. - [Destroy this (RNA) message after reading it!].
Camier S, Séraphin B. Camier S, et al. Med Sci (Paris). 2007 Oct;23(10):850-6. doi: 10.1051/medsci/20072310850. Med Sci (Paris). 2007. PMID: 17937894 Review. French.
Cited by
- NMD: a multifaceted response to premature translational termination.
Kervestin S, Jacobson A. Kervestin S, et al. Nat Rev Mol Cell Biol. 2012 Nov;13(11):700-12. doi: 10.1038/nrm3454. Epub 2012 Oct 17. Nat Rev Mol Cell Biol. 2012. PMID: 23072888 Free PMC article. Review. - Implications of targeted genomic disruption of β-catenin in BxPC-3 pancreatic adenocarcinoma cells.
Olsen PA, Solberg NT, Lund K, Vehus T, Gelazauskaite M, Wilson SR, Krauss S. Olsen PA, et al. PLoS One. 2014 Dec 23;9(12):e115496. doi: 10.1371/journal.pone.0115496. eCollection 2014. PLoS One. 2014. PMID: 25536063 Free PMC article. - Exosomal Protein Deficiencies: How Abnormal RNA Metabolism Results in Childhood-Onset Neurological Diseases.
Müller JS, Giunta M, Horvath R. Müller JS, et al. J Neuromuscul Dis. 2015;2(Suppl 2):S31-S37. doi: 10.3233/JND-150086. Epub 2015 Jul 22. J Neuromuscul Dis. 2015. PMID: 27127732 Free PMC article. - Absolute quantitative measurement of transcriptional kinetic parameters in vivo.
Iyer S, Park BR, Kim M. Iyer S, et al. Nucleic Acids Res. 2016 Oct 14;44(18):e142. doi: 10.1093/nar/gkw596. Epub 2016 Jul 4. Nucleic Acids Res. 2016. PMID: 27378780 Free PMC article. - NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure.
Eom T, Zhang C, Wang H, Lay K, Fak J, Noebels JL, Darnell RB. Eom T, et al. Elife. 2013 Jan 22;2:e00178. doi: 10.7554/eLife.00178. Elife. 2013. PMID: 23359859 Free PMC article.
References
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. - PubMed
- Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA. 2010;1:253–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM038277/GM/NIGMS NIH HHS/United States
- R01 GM074593/GM/NIGMS NIH HHS/United States
- R01 GM079707/GM/NIGMS NIH HHS/United States
- R01 GM059614/GM/NIGMS NIH HHS/United States
- R01 GM084177/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous