MiR-3120 is a mirror microRNA that targets heat shock cognate protein 70 and auxilin messenger RNAs and regulates clathrin vesicle uncoating - PubMed (original) (raw)
MiR-3120 is a mirror microRNA that targets heat shock cognate protein 70 and auxilin messenger RNAs and regulates clathrin vesicle uncoating
Helen Scott et al. J Biol Chem. 2012.
Abstract
We show that a single gene locus gives rise to two fully processed and functional miRNAs, i.e. that due to imperfect base pairing, two distinct microRNAs (miRNAs) can be produced from the fully complementary DNA strands. The antisense strand encodes miR-214, which is transcribed by its own promoter, whereas a novel miRNA, miR-3120, is co-expressed with its host gene mRNA. We also found that miR-3120 regulates important aspects of cellular function that are similar to that of its host gene, dynamin-3. miR-3120 was found to be located in neuronal cell bodies and to target Hsc70 and auxilin, and its lentivirus-mediated expression inhibited the uncoating of clathrin-coated vesicles. Finally, mirror miRNAs are likely to represent a new group of miRNAs with complex roles in coordinating gene expression.
Figures
FIGURE 1.
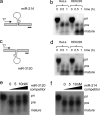
Schematic of miR-214 and miR-3120 mirror miRNAs. a, the location of the mirror miRNAs, miR-3120 and miR-214, on opposite strands of the dynamin-3 intron. b, secondary structures of the miR-3120 and miR-214 pri-miRNAs as predicted by mfold showing Drosha and dicer cleavage sites (arrows). c and d, predicted sequences of the mature 5p and 3p miRNAs (complementary sequences are starred, and mismatches are in gray).
FIGURE 2.
pri-miR-3120 is transcribed, and a mature miRNA is expressed. a and c, miR-214 (a) and miR-3120 (c) were cloned into T7 constructs, and the pri-miRNAs were produced by in vitro transcription. b and d, incubation of transcribed miR-214 (b) and miR-3120 (d) with HeLa and HEK293T cell extracts show that both miRNAs are fully processed. Note the increase in the mature miRNAs over time. e, competition assays show that the processing of miR-3120 (5 n
m
) is inhibited (as seen by the accumulation of the pri-miR-3120) by the addition of cold (0, 5, 10 n
m
) pri-miR-3120. pre, precursor-miRNA. f, processing of 32P-miR-3120 was not affected by incubation with cold pri-miR-214. The first lanes in e and f are pri-miRNAs minus HeLa cell extract.
FIGURE 3.
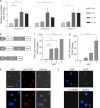
miR-3120 and miR-214 expression in neurons. a, quantitative PCR measurements in cultured neurons showing levels of endogenous miR-3120, miR-214, and dynamin-3 mRNA relative to 3 DIV. b, schematic diagram of the lentiviral constructs used. pri-miR sequences were cloned 5′ of the EGFP coding sequence and are processed out of the mRNA to produce mature miRNAs. c and d, quantitative PCR assays show that lentiviral vectors mediate an increase in miR-3120 (c) and miR-214 (d) in cultured neurons relative to nontransduced cells. MOI, multiplicity of infection. e and f, miRNAs in hippocampal neurons were visualized by in situ hybridization. DAPI staining shows nuclei in the cells (scale bar = 50 μm). Expression of endogenous miR-3120 is localized predominantly within nuclei and cytoplasm (e, top panel), and virally overexpressed miR-3120 is found to have the same distribution (e, bottom panel). The scrambled (sc-miRNA) miR-3120 probe (f, bottom panel) did not detect miR-3120 despite overexpression of miR-3120 by lentiviral vectors (confirmed as they co-express EGFP). All experiments were conducted in triplicate, and values are means ± S.E. Statistical analyses were conducted by analysis of variance and post hoc Bonferroni's test, ***, p < 0.001, **, p < 0.01, *, p < 0.05.
FIGURE 4.
miR-3120 targets conserved sites within rat and human Hsc70 UTR. a, schematic diagram of the rat and human Hsc70 3′-UTR showing species alignment and the predicted location of miR-3120 recognition sites. Black lines indicate miR-3120-3p binding, and gray lines indicate miR-3120-5p binding sites; note that the binding sites contain conserved ARE (TTTAAA) motifs. b, 3′-UTR of rat Hsc70 mRNA was co-expressed with miR-3120, control miRNA (Cont-miR) (miR-214), or an EGFP control plasmid. RLU, relative luciferase units. c, luciferase expression from reporter constructs expressing the human Hsc70 3′-UTR. d, cells transfected with an miR-3120 sponge had significantly increased Hsp70 levels. A representative Western blot is shown, and the graph represents analysis of all experiments. α-Tubulin is used as a loading control. Experiments were conducted in triplicate, and values are means ± S.E. Statistical analyses were conducted by analysis of variance and post hoc Bonferroni's test, ***, p < 0.001. Analyses were also carried out by Student's t test **, p < 0.01.
FIGURE 5.
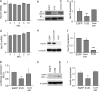
Analysis of Hsc70 and auxilin expression following transduction of miR-3120. a and d, Hsc70 mRNA levels were measured in hippocampal (a) and cortical (d) neurons following the lentivirus-mediated expression of miR-3120. MOI, multiplicity of infection. b and e, Hsc70 protein in hippocampal (b) and cortical (e) neurons following transduction with lentiviral vectors expressing miR-3120 and EGFP and miRNA controls. g–i, expression of miR-3120 also mediated a reduction in luciferase activity in auxilin 3′-UTR assays (g) and in auxilin protein expression (h). RLU, relative luciferase units. c, f, and i, densitometry analysis showed a significant reduction in hippocampal (c) and cortical (f) Hsc70 protein levels in the presence of miR-3120 and a significant reduction in auxilin levels (i). Cont-miR, control miRNA. All experiments were conducted in triplicate, and values are means ± S.E. Statistical analyses were conducted by analysis of variance and post hoc Bonferroni's test, ***, p < 0.001, **, p < 0.01, *, p < 0.05.
FIGURE 6.
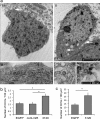
miR-3120 prevents uncoating of clathrin-coated vesicles. a, transmission electron microscopy image of EGFP transduced cell bodies (images i and ii) identified by the presence of a nucleus and surrounding cytoplasm (1,900 times end magnification). White arrow in image i denotes the pit shown in the (13,000 times) high magnification image (image iii), whereas the arrows in image ii show (13,000 times) high power images (images iv and v) of clathrin-coated vesicles. Image iii also shows ribosomes on the endoplasmic reticulum. b, clathrin-coated vesicle (CCV) counts in cortical neurons transduced with miR-3120 when compared with those transduced with EGFP and the miRNA control (Cont-miR). A similar increase in clathrin-coated vesicle number was seen in hippocampal neurons transduced with miR-3120. A detailed description of the procedures for counting clathrin-coated vesicles is given under “Results.” For both hippocampal and cortical neuronal cultures, a minimum total of 17 cells was analyzed in three individual experiments. Values are means ± S.E. Statistical analyses were conducted by analysis of variance and post hoc Bonferroni's test * = p > 0.01.
Similar articles
- Auxilin-dynamin interactions link the uncoating ATPase chaperone machinery with vesicle formation.
Newmyer SL, Christensen A, Sever S. Newmyer SL, et al. Dev Cell. 2003 Jun;4(6):929-40. doi: 10.1016/s1534-5807(03)00157-6. Dev Cell. 2003. PMID: 12791276 - Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles.
Gruschus JM, Han CJ, Greener T, Ferretti JA, Greene LE, Eisenberg E. Gruschus JM, et al. Biochemistry. 2004 Mar 23;43(11):3111-9. doi: 10.1021/bi0354740. Biochemistry. 2004. PMID: 15023062 - A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating.
Massol RH, Boll W, Griffin AM, Kirchhausen T. Massol RH, et al. Proc Natl Acad Sci U S A. 2006 Jul 5;103(27):10265-10270. doi: 10.1073/pnas.0603369103. Epub 2006 Jun 23. Proc Natl Acad Sci U S A. 2006. PMID: 16798879 Free PMC article. - Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis.
Eisenberg E, Greene LE. Eisenberg E, et al. Traffic. 2007 Jun;8(6):640-6. doi: 10.1111/j.1600-0854.2007.00568.x. Epub 2007 May 4. Traffic. 2007. PMID: 17488288 Review. - Molecular mechanism and physiological functions of clathrin-mediated endocytosis.
McMahon HT, Boucrot E. McMahon HT, et al. Nat Rev Mol Cell Biol. 2011 Jul 22;12(8):517-33. doi: 10.1038/nrm3151. Nat Rev Mol Cell Biol. 2011. PMID: 21779028 Review.
Cited by
- Non-coding RNAs turn up the heat: an emerging layer of novel regulators in the mammalian heat shock response.
Place RF, Noonan EJ. Place RF, et al. Cell Stress Chaperones. 2014 Mar;19(2):159-72. doi: 10.1007/s12192-013-0456-5. Epub 2013 Sep 4. Cell Stress Chaperones. 2014. PMID: 24002685 Free PMC article. Review. - Distinct functions of dynamin isoforms in tumorigenesis and their potential as therapeutic targets in cancer.
Meng J. Meng J. Oncotarget. 2017 Jun 20;8(25):41701-41716. doi: 10.18632/oncotarget.16678. Oncotarget. 2017. PMID: 28402939 Free PMC article. Review. - shRNA-mediated PPARα knockdown in human glioma stem cells reduces in vitro proliferation and inhibits orthotopic xenograft tumour growth.
Haynes HR, Scott HL, Killick-Cole CL, Shaw G, Brend T, Hares KM, Redondo J, Kemp KC, Ballesteros LS, Herman A, Cordero-Llana O, Singleton WG, Mills F, Batstone T, Bulstrode H, Kauppinen RA, Wurdak H, Uney JB, Short SC, Wilkins A, Kurian KM. Haynes HR, et al. J Pathol. 2019 Apr;247(4):422-434. doi: 10.1002/path.5201. Epub 2018 Dec 27. J Pathol. 2019. PMID: 30565681 Free PMC article. - A dual druggable genome-wide siRNA and compound library screening approach identifies modulators of parkin recruitment to mitochondria.
Scott HL, Buckner N, Fernandez-Albert F, Pedone E, Postiglione L, Shi G, Allen N, Wong LF, Magini L, Marucci L, O'Sullivan GA, Cole S, Powell J, Maycox P, Uney JB. Scott HL, et al. J Biol Chem. 2020 Mar 6;295(10):3285-3300. doi: 10.1074/jbc.RA119.009699. Epub 2020 Jan 7. J Biol Chem. 2020. PMID: 31911436 Free PMC article. - Evolution of the miR199-214 cluster and vertebrate skeletal development.
Desvignes T, Contreras A, Postlethwait JH. Desvignes T, et al. RNA Biol. 2014;11(4):281-94. doi: 10.4161/rna.28141. Epub 2014 Feb 20. RNA Biol. 2014. PMID: 24643020 Free PMC article.
References
- Hannon G. J. (2002) RNA interference. Nature 418, 244–251 - PubMed
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 - PubMed
- Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., Khudayberdiev S., Leuschner P. F., Busch C. J., Kane C., Hübel K., Dekker F., Hedberg C., Rengarajan B., Drepper C., Waldmann H., Kauppinen S., Greenberg M. E., Draguhn A., Rehmsmeier M., Martinez J., Schratt G. M. (2009) A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- G0700986/MRC_/Medical Research Council/United Kingdom
- MC_G0901331/MRC_/Medical Research Council/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous