Detecting and annotating genetic variations using the HugeSeq pipeline - PubMed (original) (raw)
Detecting and annotating genetic variations using the HugeSeq pipeline
Hugo Y K Lam et al. Nat Biotechnol. 2012.
No abstract available
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology.
Figures
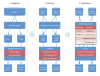
Figure 1
A MapReduce approach for detecting genetic variants from high-throughput genome sequencing. Phase 1 is the mapping phase including sequence alignment. Phase 2 is the sorting phase including sorting alignments by mapped chromosomes. Phase 3 is the reduction phase including variant detection. chr1, chromosome one; chrM, chromosome M; SV, structural variation; SR, split-read analysis; RD, read-depth analysis; RP, read-pair mapping; JM, junction mapping; VCF, variant call format; GFF, general feature format.
Figure 2
Accuracy and sensitivity of variant detection. (a) Concordant and specific calls in SNP detection by GATK and SAMtools. Original calls, total number of calls from methods; specific calls, total number of method-specific calls. Validated calls, number of calls validated by the array. Ti/Tv, transition-to-transversion rate. (b) Overall sensitivity of SNP and structural variation or CNV detection over different sequencing coverage. X is the average number of reads representing a given nucleotide in a haploid human genome. 50% indicates that detected structural variation calls overlap ≥50% the array calls.
Similar articles
- Human genome sequences: enigmatic variations.
Dunham I. Dunham I. Mutagenesis. 2002 Nov;17(6):457-61. doi: 10.1093/mutage/17.6.457. Mutagenesis. 2002. PMID: 12435842 Review. - An integrated database-pipeline system for studying single nucleotide polymorphisms and diseases.
Yang JO, Hwang S, Oh J, Bhak J, Sohn TK. Yang JO, et al. BMC Bioinformatics. 2008 Dec 12;9 Suppl 12(Suppl 12):S19. doi: 10.1186/1471-2105-9-S12-S19. BMC Bioinformatics. 2008. PMID: 19091018 Free PMC article. - An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data.
Jun G, Wing MK, Abecasis GR, Kang HM. Jun G, et al. Genome Res. 2015 Jun;25(6):918-25. doi: 10.1101/gr.176552.114. Epub 2015 Apr 16. Genome Res. 2015. PMID: 25883319 Free PMC article. - Strategies for excluding false Y-chromosomal SNP entries from human genome databases.
Rothe J, Nagy M. Rothe J, et al. Electrophoresis. 2012 May;33(9-10):1488-91. doi: 10.1002/elps.201100685. Electrophoresis. 2012. PMID: 22648820 - Current analysis platforms and methods for detecting copy number variation.
Li W, Olivier M. Li W, et al. Physiol Genomics. 2013 Jan 7;45(1):1-16. doi: 10.1152/physiolgenomics.00082.2012. Epub 2012 Nov 6. Physiol Genomics. 2013. PMID: 23132758 Free PMC article. Review.
Cited by
- Mutation spectrum in human colorectal cancers and potential functional relevance.
Yin H, Liang Y, Yan Z, Liu B, Su Q. Yin H, et al. BMC Med Genet. 2013 Mar 8;14:32. doi: 10.1186/1471-2350-14-32. BMC Med Genet. 2013. PMID: 23497483 Free PMC article. - Structural variation in the sequencing era.
Ho SS, Urban AE, Mills RE. Ho SS, et al. Nat Rev Genet. 2020 Mar;21(3):171-189. doi: 10.1038/s41576-019-0180-9. Epub 2019 Nov 15. Nat Rev Genet. 2020. PMID: 31729472 Free PMC article. Review. - Fastq2vcf: a concise and transparent pipeline for whole-exome sequencing data analyses.
Gao X, Xu J, Starmer J. Gao X, et al. BMC Res Notes. 2015 Mar 8;8:72. doi: 10.1186/s13104-015-1027-x. BMC Res Notes. 2015. PMID: 25889517 Free PMC article. - Neo-Domestication of an Interspecific Tetraploid Helianthus annuus × Helianthus tuberous Population That Segregates for Perennial Habit.
Kantar MB, Hüber S, Herman A, Bock DG, Baute G, Betts K, Ott M, Brandvain Y, Wyse D, Stupar RM, Rieseberg LH. Kantar MB, et al. Genes (Basel). 2018 Aug 21;9(9):422. doi: 10.3390/genes9090422. Genes (Basel). 2018. PMID: 30134600 Free PMC article. - Whole genome sequencing-based copy number variations reveal novel pathways and targets in Alzheimer's disease.
Ming C, Wang M, Wang Q, Neff R, Wang E, Shen Q, Reddy JS, Wang X, Allen M, Ertekin-Taner N, De Jager PL, Bennett DA, Haroutunian V, Schadt E, Zhang B. Ming C, et al. Alzheimers Dement. 2022 Oct;18(10):1846-1867. doi: 10.1002/alz.12507. Epub 2021 Dec 17. Alzheimers Dement. 2022. PMID: 34918867 Free PMC article.
References
- Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. OSDI’04 Proceedings of the 6th Symposium on Operating Systems Design and Implementation; San Francisco. 2004.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources