Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils - PubMed (original) (raw)
Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils
Wei Qiang et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Wild-type, full-length (40- and 42-residue) amyloid β-peptide (Aβ) fibrils have been shown by a variety of magnetic resonance techniques to contain cross-β structures in which the β-sheets have an in-register parallel supramolecular organization. In contrast, recent studies of fibrils formed in vitro by the Asp23-to-Asn mutant of 40-residue Aβ (D23N-Aβ(1-40)), which is associated with early onset neurodegeneration, indicate that D23N-Aβ(1-40) fibrils can contain either parallel or antiparallel β-sheets. We report a protocol for producing structurally pure antiparallel D23N-Aβ(1-40) fibril samples and a series of solid state nuclear magnetic resonance and electron microscopy measurements that lead to a specific model for the antiparallel D23N-Aβ(1-40) fibril structure. This model reveals how both parallel and antiparallel cross-β structures can be constructed from similar peptide monomer conformations and stabilized by similar sets of interactions, primarily hydrophobic in nature. We find that antiparallel D23N-Aβ(1-40) fibrils are thermodynamically metastable with respect to conversion to parallel structures, propagate less efficiently than parallel fibrils in seeded fibril growth, and therefore must nucleate more efficiently than parallel fibrils in order to be observable. Experiments in neuronal cell cultures indicate that both antiparallel and parallel D23N-Aβ(1-40) fibrils are cytotoxic. Thus, our antiparallel D23N-Aβ(1-40) fibril model represents a specific "toxic intermediate" in the aggregation process of a disease-associated Aβ mutant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
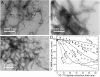
Fig. 1.
(A_–_C) TEM images of negatively stained D23N-A_β_1–40 fibrils. Images are shown for parent fibrils (A), fibrils in their thermodynamic equilibrium (TE) state after 500 h incubation with intermittent sonication (B), and fibrils prepared by two generations of the seeding/filtration (SFg2) protocol (C). The TE and SFg2 protocols select different fibril morphologies from the polymorphic parent sample. (D) Measurements of intermolecular dipole-dipole couplings among 13C labels at A21 methyl carbons in parent (squares), TE (circles) and SFg2 (triangles) fibrils, obtained with the PITHIRDS-CT SSNMR technique. Error bars represent uncertainty due to root-mean-squared noise in the 13C NMR spectra. Dashed and solid curves are simulated data for linear chains of 13C nuclei with the indicated spacings. Average intermolecular 13C-13C distances decrease in TE fibrils and increase in SFg2 fibrils, relative to distances in the parent fibrils.
Fig. 2.
(A_–_E) 2D 13C-13C correlation spectra of SFg2 fibril samples A_–_E, with color-coded chemical shift assignment paths. (F) Secondary 13C chemical shifts obtained from the 2D 13C-13C spectra. Shaded areas highlight residues with non-β-strand secondary shifts.
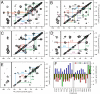
Fig. 3.
(A, B) 2D CHHC spectra of SFg2 fibril samples A and B, with 1D slices through  chemical shifts of I31 and V18 (dashed lines). Strong I31 C_α_/M35 C α and V18 C_α_/F20 C_α_ crosspeaks indicate antiparallel β-sheets. (C) 2D RAD spectrum of SFg2 fibril sample C with 1D slices at the I32
chemical shifts of I31 and V18 (dashed lines). Strong I31 C_α_/M35 C α and V18 C_α_/F20 C_α_ crosspeaks indicate antiparallel β-sheets. (C) 2D RAD spectrum of SFg2 fibril sample C with 1D slices at the I32  , L34
, L34  , and A21
, and A21  chemical shifts (dashed lines). Arrows indicate interresidue crosspeaks.
chemical shifts (dashed lines). Arrows indicate interresidue crosspeaks.
Fig. 4.
Structure of residues 15–40 in antiparallel D23N-A_β_1–40 (SFg2) fibrils. (A) Backbone atoms of the full eight-molecule system used for structure development, viewed parallel (Top) and perpendicular (Bottom) to the fibril axis. Carbon atoms are colored orange or cyan in molecules with alternating orientations within the antiparallel cross-β motif. This is one example of 10 similar structures determined from the experimental restraints (PDB file 2LNQ). See
Fig. S6
for superpositions of the 10 structures. (B) Central pair of molecules, showing all non-hydrogen atoms and viewed parallel to the fibril axis. (C) Schematic representation of the double-layered antiparallel cross-β motif (Left), showing β-strands in yellow, the intervening loop in red, and groups of hydrophobic side chains as light green and dark green blocks. A schematic representation of the double-layered parallel cross-β motif identified in earlier studies of WT-A_β_1–40 fibrils (Right) is shown for comparison.
Similar articles
- Molecular structures of amyloid and prion fibrils: consensus versus controversy.
Tycko R, Wickner RB. Tycko R, et al. Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review. - Evidence for novel beta-sheet structures in Iowa mutant beta-amyloid fibrils.
Tycko R, Sciarretta KL, Orgel JP, Meredith SC. Tycko R, et al. Biochemistry. 2009 Jul 7;48(26):6072-84. doi: 10.1021/bi9002666. Biochemistry. 2009. PMID: 19358576 Free PMC article. - Structural evolution of Iowa mutant β-amyloid fibrils from polymorphic to homogeneous states under repeated seeded growth.
Qiang W, Yau WM, Tycko R. Qiang W, et al. J Am Chem Soc. 2011 Mar 23;133(11):4018-29. doi: 10.1021/ja109679q. Epub 2011 Feb 28. J Am Chem Soc. 2011. PMID: 21355554 Free PMC article. - Supramolecular structural constraints on Alzheimer's beta-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance.
Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Antzutkin ON, et al. Biochemistry. 2002 Dec 24;41(51):15436-50. doi: 10.1021/bi0204185. Biochemistry. 2002. PMID: 12484785 - Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.
Kreutzer AG, Nowick JS. Kreutzer AG, et al. Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review.
Cited by
- Out-of-register β-sheets suggest a pathway to toxic amyloid aggregates.
Liu C, Zhao M, Jiang L, Cheng PN, Park J, Sawaya MR, Pensalfini A, Gou D, Berk AJ, Glabe CG, Nowick J, Eisenberg D. Liu C, et al. Proc Natl Acad Sci U S A. 2012 Dec 18;109(51):20913-8. doi: 10.1073/pnas.1218792109. Epub 2012 Dec 3. Proc Natl Acad Sci U S A. 2012. PMID: 23213214 Free PMC article. - Pulsed Hydrogen-Deuterium Exchange Reveals Altered Structures and Mechanisms in the Aggregation of Familial Alzheimer's Disease Mutants.
Illes-Toth E, Meisl G, Rempel DL, Knowles TPJ, Gross ML. Illes-Toth E, et al. ACS Chem Neurosci. 2021 Jun 2;12(11):1972-1982. doi: 10.1021/acschemneuro.1c00072. Epub 2021 May 14. ACS Chem Neurosci. 2021. PMID: 33988976 Free PMC article. - X-ray Crystallography Reveals Parallel and Antiparallel β-Sheet Dimers of a β-Hairpin Derived from Aβ16-36 that Assemble to Form Different Tetramers.
Kreutzer AG, Samdin TD, Guaglianone G, Spencer RK, Nowick JS. Kreutzer AG, et al. ACS Chem Neurosci. 2020 Aug 5;11(15):2340-2347. doi: 10.1021/acschemneuro.0c00290. Epub 2020 Jul 14. ACS Chem Neurosci. 2020. PMID: 32584538 Free PMC article. - General Principles Underpinning Amyloid Structure.
Taylor AIP, Staniforth RA. Taylor AIP, et al. Front Neurosci. 2022 Jun 2;16:878869. doi: 10.3389/fnins.2022.878869. eCollection 2022. Front Neurosci. 2022. PMID: 35720732 Free PMC article. Review. - A protein self-assembly model guided by electrostatic and hydrophobic dipole moments.
Mozo-Villarías A, Querol E. Mozo-Villarías A, et al. PLoS One. 2019 Apr 29;14(4):e0216253. doi: 10.1371/journal.pone.0216253. eCollection 2019. PLoS One. 2019. PMID: 31034513 Free PMC article.
References
- Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Structural features of the Aβ amyloid fibril elucidated by limited proteolysis. Biochemistry. 2001;40:11757–11767. - PubMed
- Chimon S, et al. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat Struct Mol Biol. 2007;14:1157–1164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources