Structure-based model of allostery predicts coupling between distant sites - PubMed (original) (raw)
Structure-based model of allostery predicts coupling between distant sites
Patrick Weinkam et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Allostery is a phenomenon that couples effector ligand binding at an allosteric site to a structural and/or dynamic change at a distant regulated site. To study an allosteric transition, we vary the size of the allosteric site and its interactions to construct a series of energy landscapes with pronounced minima corresponding to both the effector bound and unbound crystal structures. We use molecular dynamics to sample these landscapes. The degree of perturbation by the effector, modeled by the size of the allosteric site, provides an order parameter for allostery that allows us to determine how microscopic motions give rise to commonly discussed macroscopic mechanisms: (i) induced fit, (ii) population shift, and (iii) entropy driven. These mechanisms involve decreasing structural differences between the effector bound and unbound populations. A metric (ligand-induced cooperativity) can measure how cooperatively a given regulated site responds to effector binding and therefore what kind of allosteric mechanism is involved. We apply the model to three proteins with experimentally characterized transitions: (i) calmodulin-GFP Ca(2+) sensor protein, (ii) maltose binding protein, and (iii) CSL transcription factor. Remarkably, the model is able to reproduce allosteric motion and predict coupling in a manner consistent with experiment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
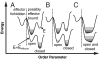
General landscapes of allostery. In an approximation, energy landscapes can be projected onto an “order parameter” that separates conformations of the system based on the structure of the regulated site. There are two landscapes pertaining to the effector bound (E + _E_effector) and unbound (E) states. Within each state there is an open substate, which occurs if the regulated site configuration is closer to that in the effector unbound crystal structure than to that of the effector bound crystal structure, and a closed substate, which occurs otherwise. The horizontal lines indicate different populated structures in each basin. Different proteins may have dissimilar landscapes, in terms of the relative heights of the barriers and basins as well as the entropy within each basin. There are three general scenarios: (A) induced fit, (B) population shift, and (C) entropy driven. For allosteric proteins, conformations not consistent with effector binding (left of the dashed line) must be less stable than bound conformations (right of the dashed line).
Fig. 2.
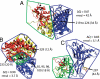
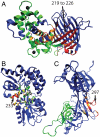
Allosteric and regulated sites. The crystal structures for (A) CaGFP, (B) MBP, and (C) CSL. The parts of the effector bound and unbound structures that differ from each other are shown in red and yellow, respectively. The effector ligand is shown in black. A radius around the effector ligand (r_AS) defines the allosteric site (green). The regulated region is shown in blue. Also shown for each structure are the distance(s) between the regulated and allosteric sites, the C_α rmsd between the bound and unbound crystal structures, and the similarity measure Δ_Q_ between the bound and unbound crystal structures. The regulated site for CaGFP is a stretch of the sequence responsible for fluorescence (residues 219–226).
Fig. 3.
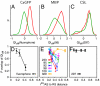
Coupling of distant sites. (A_–_C) The probability distributions of _QI_diff for the regulated sites corresponding to a large _r_AS (approximately half the distance between allosteric and regulated sites) for the effector bound (red, _QI_diff > 0) and effector unbound (green, _QI_diff < 0) simulations. (D_–_F) The _P_overlap (overlapping area between the distributions) is shown as a function of _r_AS, normalized by the distance between each regulated site and the allosteric site. The regulated sites experimentally demonstrated to be highly coupled to effector binding in solution are shown as lines with closed circles and other sites are shown as lines with open squares. Error bars represent the standard deviation calculated by randomly dividing the set of simulations into thirds.
Fig. 4.
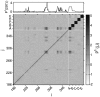
Pseudocorrelation map. A pseudocorrelation map [PC_t_-(j,i)] for the allosteric site (AS), regulated site (RS), and C-terminal (CT) domains of CSL is obtained by assigning all residues (or subsets of residues) into the effector bound or effector unbound substate using _QI_diff. (Upper) The row corresponding to the regulated site, for PC_t_-(297, i). A_–_C represent pseudocorrelations of single domains: _Q_diff (RS), _Q_diff (AS), and _Q_diff(CT), respectively. D and E represent pseudocorrelations for contacts at the interface between domains, _QI_diff(AS to RS) and _QI_diff (CT to RS), respectively.
Fig. 5.
Allosteric networks. The allosteric networks are shown for (A) CaGFP, (B) MBP, and (C) CSL. Residues are colored red when in contact and well correlated with the regulated site (labeled with arrows). A residue is considered correlated if PC_t_+(regulated site, i) has a value greater than two standard deviations above the mean PC_t_+ (Z score > 2). Residues colored orange and yellow are in contact and well correlated with red and orange residues, respectively. The remaining residues are either colored green if they are in the allosteric site (within the _r_AS radius) or blue if they are in the regulated region.
Fig. 6.
Allosteric mechanisms. (A) Diagram that qualitatively differentiates between allosteric mechanisms. LIC averaged over the whole protein (x axis) and LIC of the regulated site (y axis) are shown for CaGFP, MBP, and CSL. Points are shown for residues in the regulated site, which are defined by experimental studies, including six for MBP. Diagrams that show energy landscapes for a subset of residues in a protein are shown: (B) induced fit, (C) population shift, and (D) entropy-driven mechanisms. The arrows represent the equilibrium between the unbound (Left) and bound (Right) landscapes. The protein is divided in an allosteric site (green), regulated site (blue), allosteric network (red), and the rest (white). The sum of the contributions of individual interactions for the mechanisms in B_–_D results in landscapes shown in Fig. 1_A_–C, respectively. Higher LIC values across the whole protein often coincide with a large allosteric network and involve the cooperative motions of many residues between the allosteric site and the regulated site.
Similar articles
- The Role of Protein-Ligand Contacts in Allosteric Regulation of the Escherichia coli Catabolite Activator Protein.
Townsend PD, Rodgers TL, Glover LC, Korhonen HJ, Richards SA, Colwell LJ, Pohl E, Wilson MR, Hodgson DR, McLeish TC, Cann MJ. Townsend PD, et al. J Biol Chem. 2015 Sep 4;290(36):22225-35. doi: 10.1074/jbc.M115.669267. Epub 2015 Jul 16. J Biol Chem. 2015. PMID: 26187469 Free PMC article. - Correlating allostery with rigidity.
Rader AJ, Brown SM. Rader AJ, et al. Mol Biosyst. 2011 Feb;7(2):464-71. doi: 10.1039/c0mb00054j. Epub 2010 Nov 8. Mol Biosyst. 2011. PMID: 21060909 - Impact of mutations on the allosteric conformational equilibrium.
Weinkam P, Chen YC, Pons J, Sali A. Weinkam P, et al. J Mol Biol. 2013 Feb 8;425(3):647-61. doi: 10.1016/j.jmb.2012.11.041. Epub 2012 Dec 7. J Mol Biol. 2013. PMID: 23228330 Free PMC article. - Structural and energetic basis of allostery.
Hilser VJ, Wrabl JO, Motlagh HN. Hilser VJ, et al. Annu Rev Biophys. 2012;41:585-609. doi: 10.1146/annurev-biophys-050511-102319. Annu Rev Biophys. 2012. PMID: 22577828 Free PMC article. Review. - Protein dynamics explain the allosteric behaviors of hemoglobin.
Yonetani T, Laberge M. Yonetani T, et al. Biochim Biophys Acta. 2008 Sep;1784(9):1146-58. doi: 10.1016/j.bbapap.2008.04.025. Epub 2008 May 8. Biochim Biophys Acta. 2008. PMID: 18519045 Free PMC article. Review.
Cited by
- All-atom ensemble modeling to analyze small-angle x-ray scattering of glycosylated proteins.
Guttman M, Weinkam P, Sali A, Lee KK. Guttman M, et al. Structure. 2013 Mar 5;21(3):321-31. doi: 10.1016/j.str.2013.02.004. Structure. 2013. PMID: 23473666 Free PMC article. - Structural basis of GM-CSF and IL-2 sequestration by the viral decoy receptor GIF.
Felix J, Kandiah E, De Munck S, Bloch Y, van Zundert GC, Pauwels K, Dansercoer A, Novanska K, Read RJ, Bonvin AM, Vergauwen B, Verstraete K, Gutsche I, Savvides SN. Felix J, et al. Nat Commun. 2016 Nov 7;7:13228. doi: 10.1038/ncomms13228. Nat Commun. 2016. PMID: 27819269 Free PMC article. - SASBDB: Towards an automatically curated and validated repository for biological scattering data.
Kikhney AG, Borges CR, Molodenskiy DS, Jeffries CM, Svergun DI. Kikhney AG, et al. Protein Sci. 2020 Jan;29(1):66-75. doi: 10.1002/pro.3731. Epub 2019 Oct 11. Protein Sci. 2020. PMID: 31576635 Free PMC article. - Enhanced amino acid selection in fully evolved tryptophanyl-tRNA synthetase, relative to its urzyme, requires domain motion sensed by the D1 switch, a remote dynamic packing motif.
Weinreb V, Li L, Chandrasekaran SN, Koehl P, Delarue M, Carter CW Jr. Weinreb V, et al. J Biol Chem. 2014 Feb 14;289(7):4367-76. doi: 10.1074/jbc.M113.538660. Epub 2014 Jan 6. J Biol Chem. 2014. PMID: 24394410 Free PMC article. - Structural and Biochemical Characterization of Aldehyde Dehydrogenase 12, the Last Enzyme of Proline Catabolism in Plants.
Korasick DA, Končitíková R, Kopečná M, Hájková E, Vigouroux A, Moréra S, Becker DF, Šebela M, Tanner JJ, Kopečný D. Korasick DA, et al. J Mol Biol. 2019 Feb 1;431(3):576-592. doi: 10.1016/j.jmb.2018.12.010. Epub 2018 Dec 21. J Mol Biol. 2019. PMID: 30580036 Free PMC article.
References
- Gunasekaran K, Ma BY, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. - PubMed
- Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. - PubMed
- Koshland DE, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–368. - PubMed
- Wolynes PG. Recent successes of the energy landscape theory of protein folding and function. Q Rev Biophys. 2005;38:405–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous