Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability - PubMed (original) (raw)
. 2012 Mar 9;90(3):565-72.
doi: 10.1016/j.ajhg.2012.02.007.
Arif B Ekici, Sabine Endele, Bernt Popp, Christiane Zweier, Antje Wiesener, Eva Wohlleber, Andreas Dufke, Eva Rossier, Corinna Petsch, Markus Zweier, Ina Göhring, Alexander M Zink, Gudrun Rappold, Evelin Schröck, Dagmar Wieczorek, Olaf Riess, Hartmut Engels, Anita Rauch, André Reis
Affiliations
- PMID: 22405089
- PMCID: PMC3309205
- DOI: 10.1016/j.ajhg.2012.02.007
Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability
Juliane Hoyer et al. Am J Hum Genet. 2012.
Abstract
Intellectual disability (ID) is a clinically and genetically heterogeneous common condition that remains etiologically unresolved in the majority of cases. Although several hundred diseased genes have been identified in X-linked, autosomal-recessive, or syndromic types of ID, the establishment of an etiological basis remains a difficult task in unspecific, sporadic cases. Just recently, de novo mutations in SYNGAP1, STXBP1, MEF2C, and GRIN2B were reported as relatively common causes of ID in such individuals. On the basis of a patient with severe ID and a 2.5 Mb microdeletion including ARID1B in chromosomal region 6q25, we performed mutational analysis in 887 unselected patients with unexplained ID. In this cohort, we found eight (0.9%) additional de novo nonsense or frameshift mutations predicted to cause haploinsufficiency. Our findings indicate that haploinsufficiency of ARID1B, a member of the SWI/SNF-A chromatin-remodeling complex, is a common cause of ID, and they add to the growing evidence that chromatin-remodeling defects are an important contributor to neurodevelopmental disorders.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
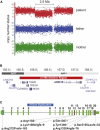
Figure 1
Summary of the Nine De Novo CNVs and Point Mutations Detected in ARID1B (A) De novo deletion in patient 1 detected with molecular karyotyping by the Affymetrix SNP6.0 platform. The signal reduction of 1,568 markers indicating the deletion region is detected only in patient 1 (red dots) and is absent in both parents. (B) 2.5 Mb deletion (red bar) in chromosomal region 6q25, which includes five RefSeq genes, among them ARID1B. Two gray scales are illustrating two different chromosomal bands as indicated in the horizontal bars. Genomic positions are in million bp. (C) Genomic structure of ARID1B. Vertical green bars illustrate the exons with their respective number above. Narrow green bars illustrate the 5′ and 3′ UTRs. The three known ARID1B domains are indicated by colored lines (blue, LXXLL; brown, ARID; and magenta, BC-Box). The duplication in patient 2 is indicated by a blue bar encompassing exons 5 and 6. The localization of mutations in patients 3–9 is indicated by black lines leading to the mutation identifiers.
Figure 2
Transcript Analysis for the ARID1B Mutations in Patients 2 and 7 (A) Partial sequence electropherograms of ARID1B exon 6 obtained from gDNA and cDNA from patient 2 with a de novo duplication of exons 5 and 6 and his healthy parents. Patient 2 and his father are heterozygous for rs3734441 (c.2172G>A on exon 6) at the gDNA level. Note that the amount of guanine is doubled in the patient. At the cDNA level, the father shows biallelic expression, whereas monoallelic expression of adenine in patient 2 indicates that the duplication leads to a null allele. (B and C) Synonymous variant in the last codon of exon 17 induces exon skipping of exon 17 in patient 7. Using primers located in exons 16 and 18 (c16f and c18r, respectively), indicated by arrows for RT-PCR on RNA from peripheral blood leukocytes, resulted in an additional aberrant product of 148 bp in the patient (lane 3), whereas both parents (lanes 1 and 2) and controls (lanes 4–6) showed only the expected 245 bp fragment (lane 7, genomic DNA control; lane 8, no template control; M, size standard). Exon skipping in the patient was verified by the sequencing of amplified products and is predicted to result in a frameshift and premature stop codon after 76 amino acids (p.Arg1338Argfs∗76). In the patient's lane, an additional third band with a molecular size of ∼310 bp represents a heteroduplex.
Figure 3
Facial Appearance of Patients with Deletions or Mutations in ARID1B Note consistently low-set, posteriorly rotated, and abnormally shaped ears and other frequent dysmorphisms such as frontal bossing, downslanting palpebral fissures, a bulbous nasal tip, and a thin upper lip. (A and B) Patient 1 at age 3 years, 3 months. (C and D) Patient 2 at age 4 years, 11 months. (E and F) Patient 3 at age 3 years, 5 months. (G) Patient 4 at age 5 months. (H and I) Patient 4 at age 7 years, 3 months. (J) Patient 5 at age 4 years, 11 months. (K and L) Patient 5 at age 12 years, 8 months. (M and N) Patient 6 at age 6 years, 3 months. (O and P) Patient 7 at age 3 years, 10 months. (Q and R) Patient 8 at age 17 years.
Similar articles
- BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription.
Dias C, Estruch SB, Graham SA, McRae J, Sawiak SJ, Hurst JA, Joss SK, Holder SE, Morton JE, Turner C, Thevenon J, Mellul K, Sánchez-Andrade G, Ibarra-Soria X, Deriziotis P, Santos RF, Lee SC, Faivre L, Kleefstra T, Liu P, Hurles ME; DDD Study; Fisher SE, Logan DW. Dias C, et al. Am J Hum Genet. 2016 Aug 4;99(2):253-74. doi: 10.1016/j.ajhg.2016.05.030. Epub 2016 Jul 21. Am J Hum Genet. 2016. PMID: 27453576 Free PMC article. - Clinical correlations of mutations affecting six components of the SWI/SNF complex: detailed description of 21 patients and a review of the literature.
Kosho T, Okamoto N, Ohashi H, Tsurusaki Y, Imai Y, Hibi-Ko Y, Kawame H, Homma T, Tanabe S, Kato M, Hiraki Y, Yamagata T, Yano S, Sakazume S, Ishii T, Nagai T, Ohta T, Niikawa N, Mizuno S, Kaname T, Naritomi K, Narumi Y, Wakui K, Fukushima Y, Miyatake S, Mizuguchi T, Saitsu H, Miyake N, Matsumoto N. Kosho T, et al. Am J Med Genet A. 2013 Jun;161A(6):1221-37. doi: 10.1002/ajmg.a.35933. Epub 2013 May 1. Am J Med Genet A. 2013. PMID: 23637025 Review. - Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome.
Santen GW, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, Kant SG, Snoeck IN, Peeters EA, Hilhorst-Hofstee Y, Wessels MW, den Hollander NS, Ruivenkamp CA, van Ommen GJ, Breuning MH, den Dunnen JT, van Haeringen A, Kriek M. Santen GW, et al. Nat Genet. 2012 Mar 18;44(4):379-80. doi: 10.1038/ng.2217. Nat Genet. 2012. PMID: 22426309 - Expanding the phenotypic spectrum of ARID1B-mediated disorders and identification of altered cell-cycle dynamics due to ARID1B haploinsufficiency.
Sim JC, White SM, Fitzpatrick E, Wilson GR, Gillies G, Pope K, Mountford HS, Torring PM, McKee S, Vulto-van Silfhout AT, Jhangiani SN, Muzny DM, Leventer RJ, Delatycki MB, Amor DJ, Lockhart PJ. Sim JC, et al. Orphanet J Rare Dis. 2014 Mar 27;9:43. doi: 10.1186/1750-1172-9-43. Orphanet J Rare Dis. 2014. PMID: 24674232 Free PMC article. - SWI/SNF complex in disorder: SWItching from malignancies to intellectual disability.
Santen GW, Kriek M, van Attikum H. Santen GW, et al. Epigenetics. 2012 Nov;7(11):1219-24. doi: 10.4161/epi.22299. Epub 2012 Sep 25. Epigenetics. 2012. PMID: 23010866 Free PMC article. Review.
Cited by
- BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription.
Dias C, Estruch SB, Graham SA, McRae J, Sawiak SJ, Hurst JA, Joss SK, Holder SE, Morton JE, Turner C, Thevenon J, Mellul K, Sánchez-Andrade G, Ibarra-Soria X, Deriziotis P, Santos RF, Lee SC, Faivre L, Kleefstra T, Liu P, Hurles ME; DDD Study; Fisher SE, Logan DW. Dias C, et al. Am J Hum Genet. 2016 Aug 4;99(2):253-74. doi: 10.1016/j.ajhg.2016.05.030. Epub 2016 Jul 21. Am J Hum Genet. 2016. PMID: 27453576 Free PMC article. - HSA21 Single-Minded 2 (Sim2) Binding Sites Co-Localize with Super-Enhancers and Pioneer Transcription Factors in Pluripotent Mouse ES Cells.
Letourneau A, Cobellis G, Fort A, Santoni F, Garieri M, Falconnet E, Ribaux P, Vannier A, Guipponi M, Carninci P, Borel C, Antonarakis SE. Letourneau A, et al. PLoS One. 2015 May 8;10(5):e0126475. doi: 10.1371/journal.pone.0126475. eCollection 2015. PLoS One. 2015. PMID: 25955728 Free PMC article. - Brief Report: SETD2 Mutation in a Child with Autism, Intellectual Disabilities and Epilepsy.
Lumish HS, Wynn J, Devinsky O, Chung WK. Lumish HS, et al. J Autism Dev Disord. 2015 Nov;45(11):3764-70. doi: 10.1007/s10803-015-2484-8. J Autism Dev Disord. 2015. PMID: 26084711 - Inability to switch from ARID1A-BAF to ARID1B-BAF impairs exit from pluripotency and commitment towards neural crest formation in ARID1B-related neurodevelopmental disorders.
Pagliaroli L, Porazzi P, Curtis AT, Scopa C, Mikkers HMM, Freund C, Daxinger L, Deliard S, Welsh SA, Offley S, Ott CA, Calabretta B, Brugmann SA, Santen GWE, Trizzino M. Pagliaroli L, et al. Nat Commun. 2021 Nov 9;12(1):6469. doi: 10.1038/s41467-021-26810-x. Nat Commun. 2021. PMID: 34753942 Free PMC article. - High frequency strand slippage mutations in CTCF in MSI-positive endometrial cancers.
Zighelboim I, Mutch DG, Knapp A, Ding L, Xie M, Cohn DE, Goodfellow PJ. Zighelboim I, et al. Hum Mutat. 2014 Jan;35(1):63-5. doi: 10.1002/humu.22463. Hum Mutat. 2014. PMID: 24130125 Free PMC article.
References
- Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. - PubMed
- Abou Jamra R., Philippe O., Raas-Rothschild A., Eck S.H., Graf E., Buchert R., Borck G., Ekici A., Brockschmidt F.F., Nöthen M.M. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am. J. Hum. Genet. 2011;88:788–795. - PMC - PubMed
- Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. - PubMed
- Endele S., Rosenberger G., Geider K., Popp B., Tamer C., Stefanova I., Milh M., Kortüm F., Fritsch A., Pientka F.K. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat. Genet. 2010;42:1021–1026. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous