Identification and characterization of a bacterial hydrosulphide ion channel - PubMed (original) (raw)
Identification and characterization of a bacterial hydrosulphide ion channel
Bryan K Czyzewski et al. Nature. 2012.
Abstract
The hydrosulphide ion (HS(-)) and its undissociated form, hydrogen sulphide (H(2)S), which are believed to have been critical to the origin of life on Earth, remain important in physiology and cellular signalling. As a major metabolite in anaerobic bacterial growth, hydrogen sulphide is a product of both assimilatory and dissimilatory sulphate reduction. These pathways can reduce various oxidized sulphur compounds including sulphate, sulphite and thiosulphate. The dissimilatory sulphate reduction pathway uses this molecule as the terminal electron acceptor for anaerobic respiration, in which process it produces excess amounts of H(2)S (ref. 4). The reduction of sulphite is a key intermediate step in all sulphate reduction pathways. In Clostridium and Salmonella, an inducible sulphite reductase is directly linked to the regeneration of NAD(+), which has been suggested to have a role in energy production and growth, as well as in the detoxification of sulphite. Above a certain concentration threshold, both H(2)S and HS(-) inhibit cell growth by binding the metal centres of enzymes and cytochrome oxidase, necessitating a release mechanism for the export of this toxic metabolite from the cell. Here we report the identification of a hydrosulphide ion channel in the pathogen Clostridium difficile through a combination of genetic, biochemical and functional approaches. The HS(-) channel is a member of the formate/nitrite transport family, in which about 50 hydrosulphide ion channels form a third subfamily alongside those for formate (FocA) and for nitrite (NirC). The hydrosulphide ion channel is permeable to formate and nitrite as well as to HS(-) ions. Such polyspecificity can be explained by the conserved ion selectivity filter observed in the channel's crystal structure. The channel has a low open probability and is tightly regulated, to avoid decoupling of the membrane proton gradient.
Conflict of interest statement
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Figures
Fig. 1
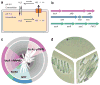
Genetic analyses and functional characterization of the FNT3/HSC gene and the asrABC operon. a, Model of the intracellular anion concentrative effect for the weak acid H2S. b, Genomic organization of FNT and their metabolically related reductase genes. Representatives are shown for the focA, nirC and FNT3/HSC genes with their respectively linked operons. c, Phylogentic tree of 474 bacterial and archaeal members of the FNT family. Branches are colorized based on genetic linkage to metabolic enzymes: pyruvate formate lyase (pflAB) or formate dehydrogenase (fdhAB) linked genes are colored pink, nitrite reductase (nirBD) linked genes are colored blue, and sulfite reductase linked genes (asrABC) are colored green. The FocA protein in archaea is encoded by the fdhC gene. The gray areas represent FNT family members with no assigned function based on genetic linkage. d, Bismuth sulfite agar plate assay. Top: vector control, left: asrA, asrB, asrC, right: asrA, asrB, asrC, FNT3.
Fig 2
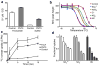
Binding and transport activity of HSC channel in reconstituted proteoliposomes. a, Measurements of sulfide concentrations in the media of salmonella transformed with vector control or vector encoding FNT3. Minimal media was supplemented with either sulfite or thiosulfate to induce hydrogen sulfide production from either periplasmic thiosulfate reductase or cytoplasmic sulfite reductase. b, Binding of detergent solubilized and purified HSC protein to various anions was determined by using thermostability coupled size exclusion chromatography. Peak heights of recovered samples were plotted against temperature and fitted to a boltzman-sigmoidal model to determine nominal melting temperatures. c, Radiolabeled formate uptake in proteoliposomes reconstituted with purified FNT at pH 8.0 was monitored in a concentrative uptake assay and compared to FocA activity or vesicle controls. d, Inhibition of radiolabeled concentrative uptake of formate by the addition of various anions at increasing concentrations. The plotted bar graphs represent the amount of radiolabeled formate measured at the 10-minute time point for each concentration of anion tested. The concentration of the competing anions were, 0 mM, 0.15 mM, 0.6 mM, 3 mM and 15 mM. Error bars represent s.e.m. (N = 3).
Fig 3
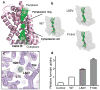
Structural and functional characterization of the ion permeation pathway. a, Structure of a protomer of HSC overlaid with the pore diameter calculations from HOLE, colorized to indicate the radius of water, where green is permeable to water and red is impermeable. Transmembrane helix-2 has been removed for clarity. b, Pore diameter calculations from HOLE of the crystal structures for each of the two permeation pathway mutations, Leu82Val and Phe194Ile and wild-type (WT). c, Close up of the electron density observed at the selectivity filter for the Leu82Val mutant. Purple is the 2Fo – Fc electron density map contoured at 1.1σ and green is the Fo – Fc difference density map contoured at 3σ. d, Relative uptake of proteoliposomes reconstituted with purified HSC protein and pathway mutations. The bar graph represents the 10-minute time point of each concentrative uptake experiment. Error bars represent s.e.m. (N = 3).
Fig 4
Structural and functional characterization of possible gating mechanisms. a, Structure of the HSC protomer representing possible gating regions. The structure is colorized to show the two-fold inverted topology. b&c, The Glu-Lys-Asn salt bridge triads, related by a pseudo-twofold symmetry, help to stabilize the helix-P at the periplasmic side and helix-N at the cytoplasmic side of the protein. The rotamer that each residue adopts is conserved. d, Relative uptake of proteoliposomes reconstituted with purified HSC protein and gating mutations. The bar graph represents the 10-minute time point of each concentrative uptake experiment. Error bars represent s.e.m. (N = 3).
Similar articles
- Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium.
Lü W, Schwarzer NJ, Du J, Gerbig-Smentek E, Andrade SL, Einsle O. Lü W, et al. Proc Natl Acad Sci U S A. 2012 Nov 6;109(45):18395-400. doi: 10.1073/pnas.1210793109. Epub 2012 Oct 22. Proc Natl Acad Sci U S A. 2012. PMID: 23090993 Free PMC article. - pH-dependent gating in a FocA formate channel.
Lü W, Du J, Wacker T, Gerbig-Smentek E, Andrade SL, Einsle O. Lü W, et al. Science. 2011 Apr 15;332(6027):352-4. doi: 10.1126/science.1199098. Science. 2011. PMID: 21493860 - The formate channel FocA exports the products of mixed-acid fermentation.
Lü W, Du J, Schwarzer NJ, Gerbig-Smentek E, Einsle O, Andrade SL. Lü W, et al. Proc Natl Acad Sci U S A. 2012 Aug 14;109(33):13254-9. doi: 10.1073/pnas.1204201109. Epub 2012 Jul 30. Proc Natl Acad Sci U S A. 2012. PMID: 22847446 Free PMC article. - The formate/nitrite transporter family of anion channels.
Lü W, Du J, Schwarzer NJ, Wacker T, Andrade SL, Einsle O. Lü W, et al. Biol Chem. 2013 Jun;394(6):715-27. doi: 10.1515/hsz-2012-0339. Biol Chem. 2013. PMID: 23380538 Review. - Voltage-gated proton channels and other proton transfer pathways.
Decoursey TE. Decoursey TE. Physiol Rev. 2003 Apr;83(2):475-579. doi: 10.1152/physrev.00028.2002. Physiol Rev. 2003. PMID: 12663866 Review.
Cited by
- Hydrogen sulfide and polysulfides as signaling molecules.
Kimura H. Kimura H. Proc Jpn Acad Ser B Phys Biol Sci. 2015;91(4):131-59. doi: 10.2183/pjab.91.131. Proc Jpn Acad Ser B Phys Biol Sci. 2015. PMID: 25864468 Free PMC article. Review. - Disentangling mechanisms involved in the adaptation of photosynthetic microorganisms to the extreme sulphureous water from Los Baños de Vilo (S Spain).
del Mar Fernández-Arjona M, Bañares-España E, García-Sánchez MJ, Hernández-López M, López-Rodas V, Costas E, Flores-Moya A. del Mar Fernández-Arjona M, et al. Microb Ecol. 2013 Nov;66(4):742-51. doi: 10.1007/s00248-013-0268-2. Epub 2013 Jul 24. Microb Ecol. 2013. PMID: 23880793 - Regulation of Ion Channel Function by Gas Molecules.
Shah N, Zhou L. Shah N, et al. Adv Exp Med Biol. 2021;1349:139-164. doi: 10.1007/978-981-16-4254-8_8. Adv Exp Med Biol. 2021. PMID: 35138614 Review. - A single amino acid exchange converts FocA into a unidirectional efflux channel for formate.
Kammel M, Trebbin O, Pinske C, Sawers RG. Kammel M, et al. Microbiology (Reading). 2022 Jan;168(1):001132. doi: 10.1099/mic.0.001132. Microbiology (Reading). 2022. PMID: 35084298 Free PMC article. - The C-terminal Six Amino Acids of the FNT Channel FocA Are Required for Formate Translocation But Not Homopentamer Integrity.
Hunger D, Röcker M, Falke D, Lilie H, Sawers RG. Hunger D, et al. Front Microbiol. 2017 Aug 22;8:1616. doi: 10.3389/fmicb.2017.01616. eCollection 2017. Front Microbiol. 2017. PMID: 28878762 Free PMC article.
References
- Wachtershauser G. Groundworks for an evolutionary biochemistry: the iron-sulphur world. Prog Biophys Mol Biol. 1992;58:85–201. - PubMed
- Dhillon A, Goswami S, Riley M, Teske A, Sogin M. Domain evolution and functional diversification of sulfite reductases. Astrobiology. 2005;5:18–29. - PubMed
- Rabus R, Hansen T, Widdel F. Dissimilatory sulfate- and sulfur-reducing prokaryotes. In: Dworkin M, et al., editors. The Prokaryotes: Ecophysiology and Biochemistry. Vol. 2. Springer; Heidelberg: 2006. pp. 659–768.
- Goffredi SK, Childress JJ, Desaulniers NT, Lallier FJ. Sulfide acquisition by the vent worm Riftia pachyptila appears to be via uptake of HS−, rather than H2S. J Exp Biol. 1997;200:2609–2616. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54-GM075026/GM/NIGMS NIH HHS/United States
- R01 GM093825/GM/NIGMS NIH HHS/United States
- R01-DK073973/DK/NIDDK NIH HHS/United States
- R01 MH083840/MH/NIMH NIH HHS/United States
- R01 DK053973/DK/NIDDK NIH HHS/United States
- F31 AI086072/AI/NIAID NIH HHS/United States
- R01-MH083840/MH/NIMH NIH HHS/United States
- U54 GM075026/GM/NIGMS NIH HHS/United States
- R01-DK053973-08A1S1/DK/NIDDK NIH HHS/United States
- F31-AI086072/AI/NIAID NIH HHS/United States
- R01 DK073973/DK/NIDDK NIH HHS/United States
- R01-GM093825/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials