Common origins and host-dependent diversity of plant and animal viromes - PubMed (original) (raw)
Review
Common origins and host-dependent diversity of plant and animal viromes
Valerian V Dolja et al. Curr Opin Virol. 2011 Nov.
Abstract
Many viruses infecting animals and plants share common cores of homologous genes involved in the key processes of viral replication. In contrast, genes that mediate virus–host interactions including in many cases capsid protein (CP) genes are markedly different. There are three distinct scenarios for the origin of related viruses of plants and animals: first, evolution from a common ancestral virus predating the divergence of plants and animals; second, horizontal transfer of viruses, for example, through insect vectors; third, parallel origin from related genetic elements. We present evidence that each of these scenarios contributed, to a varying extent, to the evolution of different groups of viruses.
Figures
Figure 1
Global ecology of viruses. The shapes denoting virus taxa are color-coded in accordance with their host range as indicated in the figure. NCLDV, Nucleo-cytoplasmic large DNA viruses; SF, superfamily.
Figure 2
Comparison of genome architectures of related plant and animal viruses: The housekeeping and interactive gene modules. The virus genes are drawn as boxes approximately to scale. For each pair of animal (pink background) and plant (green background) viruses, the homologous genes are shown in the same color. Gene designations: VP, virus protein; S3H, superfamily 3 helicase; g, virus protein, genome-linked; RdRp, Rna-dependent RNA polymerase; MP, movement protein; CP, capsid protein; ProC, protease cofactor; C-Pro, cysteine protease; nsP, non-structural protein; CAP, capping enzyme; S1H, superfamily 1 helicase; P-Pro, papain-like protease; NC, nucleocapsid protein; E, envelope protein; C, nucleocapsid protein; prM, pre-membrane protein; NS, non-structural protein; Pro, protease; S2H, superfamily 2 helicase; MET; methyltransferase; G, glycoprotein; N, nucleocapsid protein; RCRE, rolling-circle replication endonuclease; NSP, nuclear shuttle protein. Virus abbreviations: PolioV, Poliovirus; CPMV, Cowpea mosaic virus; SINV, Sindbis virus; TMV, Tobacco mosaic virus; DENV Dengue fever virus; TBSV, Tomato bushy stunt virus; RVFV, Rift valley fever virus; TSWV, Tomato spotted wilt virus.
Figure 2
Comparison of genome architectures of related plant and animal viruses: The housekeeping and interactive gene modules. The virus genes are drawn as boxes approximately to scale. For each pair of animal (pink background) and plant (green background) viruses, the homologous genes are shown in the same color. Gene designations: VP, virus protein; S3H, superfamily 3 helicase; g, virus protein, genome-linked; RdRp, Rna-dependent RNA polymerase; MP, movement protein; CP, capsid protein; ProC, protease cofactor; C-Pro, cysteine protease; nsP, non-structural protein; CAP, capping enzyme; S1H, superfamily 1 helicase; P-Pro, papain-like protease; NC, nucleocapsid protein; E, envelope protein; C, nucleocapsid protein; prM, pre-membrane protein; NS, non-structural protein; Pro, protease; S2H, superfamily 2 helicase; MET; methyltransferase; G, glycoprotein; N, nucleocapsid protein; RCRE, rolling-circle replication endonuclease; NSP, nuclear shuttle protein. Virus abbreviations: PolioV, Poliovirus; CPMV, Cowpea mosaic virus; SINV, Sindbis virus; TMV, Tobacco mosaic virus; DENV Dengue fever virus; TBSV, Tomato bushy stunt virus; RVFV, Rift valley fever virus; TSWV, Tomato spotted wilt virus.
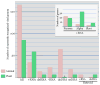
Figure 3
Prevalence of major groups of viruses in plant and animal viromes. The bar graphs show the number of virus genera infecting plants and animals for each major group of viruses. The asterisks indicates that the only known group of typical dsDNA viruses in the plant kingdom, the Phycodnaviridae, infects green algae rather than land plants. Virus taxonomy was from the NCBI Taxonomy Browser:
Figure 4
The three scenarios for the origin of related viruses in plants and animals: Common ancestry, horizontal virus transfer and parallel evolution. For each scenario, examples of virus groups to which it most likely applies are given (see text). Abbreviations: LECA, Last eukaryotic common ancestor; MP, movement protein; HVT, horizontal virus transfer; RCRE, rolling-circle replication endonuclease; CP, capsid protein.
Similar articles
- Can plant viruses cross the kingdom border and be pathogenic to humans?
Balique F, Lecoq H, Raoult D, Colson P. Balique F, et al. Viruses. 2015 Apr 20;7(4):2074-98. doi: 10.3390/v7042074. Viruses. 2015. PMID: 25903834 Free PMC article. Review. - Creation of diversity in the animal virus world by inter-species and intra-species recombinations: lessons learned from poultry viruses.
Davidson I, Silva RF. Davidson I, et al. Virus Genes. 2008 Feb;36(1):1-9. doi: 10.1007/s11262-007-0165-1. Epub 2007 Oct 6. Virus Genes. 2008. PMID: 17922180 Review. - Insect transmission of plant viruses: a constraint on virus variability.
Power AG. Power AG. Curr Opin Plant Biol. 2000 Aug;3(4):336-40. doi: 10.1016/s1369-5266(00)00090-x. Curr Opin Plant Biol. 2000. PMID: 10873852 Review. - Plant Virus Genome Is Shaped by Specific Dinucleotide Restrictions That Influence Viral Infection.
González de Prádena A, Sánchez Jimenez A, San León D, Simmonds P, García JA, Valli AA. González de Prádena A, et al. mBio. 2020 Feb 18;11(1):e02818-19. doi: 10.1128/mBio.02818-19. mBio. 2020. PMID: 32071264 Free PMC article. - Virus-induced aggregates in infected cells.
Moshe A, Gorovits R. Moshe A, et al. Viruses. 2012 Oct 17;4(10):2218-32. doi: 10.3390/v4102218. Viruses. 2012. PMID: 23202461 Free PMC article. Review.
Cited by
- Can plant viruses cross the kingdom border and be pathogenic to humans?
Balique F, Lecoq H, Raoult D, Colson P. Balique F, et al. Viruses. 2015 Apr 20;7(4):2074-98. doi: 10.3390/v7042074. Viruses. 2015. PMID: 25903834 Free PMC article. Review. - Mutation-selection balance and compensatory mechanisms in tumour evolution.
Persi E, Wolf YI, Horn D, Ruppin E, Demichelis F, Gatenby RA, Gillies RJ, Koonin EV. Persi E, et al. Nat Rev Genet. 2021 Apr;22(4):251-262. doi: 10.1038/s41576-020-00299-4. Epub 2020 Nov 30. Nat Rev Genet. 2021. PMID: 33257848 Review. - Overview on Sobemoviruses and a Proposal for the Creation of the Family Sobemoviridae.
Sõmera M, Sarmiento C, Truve E. Sõmera M, et al. Viruses. 2015 Jun 15;7(6):3076-115. doi: 10.3390/v7062761. Viruses. 2015. PMID: 26083319 Free PMC article. Review. - Plant Viral Proteases: Beyond the Role of Peptide Cutters.
Rodamilans B, Shan H, Pasin F, García JA. Rodamilans B, et al. Front Plant Sci. 2018 May 17;9:666. doi: 10.3389/fpls.2018.00666. eCollection 2018. Front Plant Sci. 2018. PMID: 29868107 Free PMC article. Review. - Phytopathogenic fungus hosts a plant virus: A naturally occurring cross-kingdom viral infection.
Andika IB, Wei S, Cao C, Salaipeth L, Kondo H, Sun L. Andika IB, et al. Proc Natl Acad Sci U S A. 2017 Nov 14;114(46):12267-12272. doi: 10.1073/pnas.1714916114. Epub 2017 Oct 30. Proc Natl Acad Sci U S A. 2017. PMID: 29087346 Free PMC article.
References
- Keeling PJ. Genomics. Deep questions in the tree of life. Science. 2007;317:1875–6. - PubMed
- Suttle CA. Marine viruses - major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. - PubMed
- Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. This overview of the results of marine virology characterizes viruses as a major biogeochemical factor that affects the environment on the one hand by killing cells and releasing their content, and on the other hand, by enabling cells to occupy new ecological niches using genes transferred by viruses. - PubMed
- Kristensen DM, Mushegian A, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. An analytic review of virus metagenomics showing how metagenomics changes the prevailing ideas on the structure of the virus world. Statistical analysis of viral metagenomic sequences suggests that the marine DNA virome could be dominated by virus-like particles encapsidating host DNA, known as Gene Transfer Agents. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous