Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome - PubMed (original) (raw)
Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome
J K Greenberg et al. Genes Brain Behav. 2012 Aug.
Abstract
Small, non-coding microRNAs (miRNAs) have been implicated in many biological processes, including the development of the nervous system. However, the roles of miRNAs in natural behavioral and neuronal plasticity are not well understood. To help address this we characterized the microRNA transcriptome in the adult worker honey bee head and investigated whether changes in microRNA expression levels in the brain are associated with division of labor among honey bees, a well-established model for socially regulated behavior. We determined that several miRNAs were downregulated in bees that specialize on brood care (nurses) relative to foragers. Additional experiments showed that this downregulation is dependent upon social context; it only occurred when nurse bees were in colonies that also contained foragers. Analyses of conservation patterns of brain-expressed miRNAs across Hymenoptera suggest a role for certain miRNAs in the evolution of the Aculeata, which includes all the eusocial hymenopteran species. Our results support the intriguing hypothesis that miRNAs are important regulators of social behavior at both developmental and evolutionary time scales.
© 2012 The Authors. Genes, Brain and Behavior © 2012 Blackwell Publishing Ltd and International Behavioural and Neural Genetics Society.
Figures
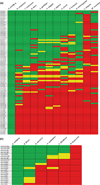
Figure 1. Conservation of miRNAs across Insecta
(a) All identified miRNAs in the honey bee head were used as probes in BLAST searches of various representative insect species, as well as to Caenorhabditis elegans, and Mus musculus, which represented non-insect outgroups. Green highlighting indicates the miRNA is conserved. Yellow highlighting indicates similarity with a single, mid-sequence base changed. Red highlighting indicates a miRNA is not present. (b) Conservation of miRNAs that appeared honey bee-specific in eusocial as well as non-social Hymenoptera. Color coding as in (a). Note that no miRNAs that appeared honey bee-specific in (a) were conserved in Nasonia longicornis, a nonsocial wasp.
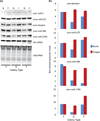
Figure 2. Relative brain expression levels of miRNAs as a function of behavioral maturation
Bees were collected from either typical (T), or young (Y) or old (O) single-cohort colonies as either nurses (N) or foragers (F). Bees from young single-cohort colonies were 7–10-days old, while bees from old single-cohort colonies were >3-weeks old. (a) Northern blot expression for five miRNAs, along with the control U6snRNA and total RNA expression. (b) Quantified miRNA expression (total area of RNA band) normalized to U6snRNA.
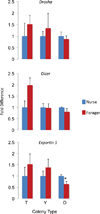
Figure 3. Relative brain mRNA expression levels of miRNA processing proteins as a function of behavioral maturation
Bees were the same as those depicted in Fig. 4 (independent samples t test for each colony). df = 6 for typical, young and old colonies). N = 4 per group. * indicates P < 0.05.
Figure 4. The ame-miR-2796 and PLC-epsilon loci
(a) Structure of ame-miR-2796 pre-miRNA with hairpin loop, mature sequence (red box), and miRNA* (blue box). (b) Alignment of the genomic region around ame-miR-2796 in five insect species. (c) Relative expression of ame-miR-2796 in honey bee brain, gland, and all other head tissues left in the carcass (Other). (d) The genomic architecture of the PLC-epsilon gene in Apis and Tribolium. Green boxes are exons and the red box (marked by arrow) shows the location of ame-miR-2796. (e) Predicted targeting sites for ame-miR-2796 in the PLC-epsilon transcripts of Apis and Tribolium. No targeting sites were identified in the PLC-epsilon 3′ UTR. Minimum free energy of hybridization is shown below each duplex prediction. (f) Relative expression of PLC-epsilon in honey bee brain, gland, and all other head tissue (Other). (g) Protein tree of PLC-epsilon. (h) Relative expression of PLC-epsilon in typical, young, and old honeybee colonies. (Independent samples t test. df = 6 for typical, young, and old colonies). N = 4 per group. * indicates P < 0.05.
Similar articles
- A role for octopamine in honey bee division of labor.
Schulz DJ, Barron AB, Robinson GE. Schulz DJ, et al. Brain Behav Evol. 2002;60(6):350-9. doi: 10.1159/000067788. Brain Behav Evol. 2002. PMID: 12563167 - Honey bee (Apis mellifera) larval pheromones may regulate gene expression related to foraging task specialization.
Ma R, Rangel J, Grozinger CM. Ma R, et al. BMC Genomics. 2019 Jul 19;20(1):592. doi: 10.1186/s12864-019-5923-7. BMC Genomics. 2019. PMID: 31324147 Free PMC article. - Biogenic amines and division of labor in honey bee colonies.
Wagener-Hulme C, Kuehn JC, Schulz DJ, Robinson GE. Wagener-Hulme C, et al. J Comp Physiol A. 1999 May;184(5):471-9. doi: 10.1007/s003590050347. J Comp Physiol A. 1999. PMID: 10377980 - Defensive behavior of honey bees: organization, genetics, and comparisons with other bees.
Breed MD, Guzmán-Novoa E, Hunt GJ. Breed MD, et al. Annu Rev Entomol. 2004;49:271-98. doi: 10.1146/annurev.ento.49.061802.123155. Annu Rev Entomol. 2004. PMID: 14651465 Review. - Juvenile hormone, behavioral maturation, and brain structure in the honey bee.
Fahrbach SE, Robinson GE. Fahrbach SE, et al. Dev Neurosci. 1996;18(1-2):102-14. doi: 10.1159/000111474. Dev Neurosci. 1996. PMID: 8840089 Review.
Cited by
- Epigenetic modification of gene expression in honey bees by heterospecific gland secretions.
Shi YY, Wu XB, Huang ZY, Wang ZL, Yan WY, Zeng ZJ. Shi YY, et al. PLoS One. 2012;7(8):e43727. doi: 10.1371/journal.pone.0043727. Epub 2012 Aug 21. PLoS One. 2012. PMID: 22928024 Free PMC article. - MicroRNAs derived from the insect virus HzNV-1 promote lytic infection by suppressing histone methylation.
Wu PC, Lin YH, Wu TC, Lee ST, Wu CP, Chang Y, Wu YL. Wu PC, et al. Sci Rep. 2018 Dec 13;8(1):17817. doi: 10.1038/s41598-018-35782-w. Sci Rep. 2018. PMID: 30546025 Free PMC article. - Physical Enrichment Triggers Brain Plasticity and Influences Blood Plasma Circulating miRNA in Rainbow Trout (Oncorhynchus mykiss).
Cardona E, Brunet V, Baranek E, Milhade L, Skiba-Cassy S, Bobe J, Calandreau L, Roy J, Colson V. Cardona E, et al. Biology (Basel). 2022 Jul 22;11(8):1093. doi: 10.3390/biology11081093. Biology (Basel). 2022. PMID: 35892949 Free PMC article. - Do social insects support Haig's kin theory for the evolution of genomic imprinting?
Pegoraro M, Marshall H, Lonsdale ZN, Mallon EB. Pegoraro M, et al. Epigenetics. 2017 Sep;12(9):725-742. doi: 10.1080/15592294.2017.1348445. Epigenetics. 2017. PMID: 28703654 Free PMC article. Review. - What do studies of insect polyphenisms tell us about nutritionally-triggered epigenomic changes and their consequences?
Cridge AG, Leask MP, Duncan EJ, Dearden PK. Cridge AG, et al. Nutrients. 2015 Mar 11;7(3):1787-97. doi: 10.3390/nu7031787. Nutrients. 2015. PMID: 25768950 Free PMC article. Review.
References
- Andersson M. The evolution of eusociality. Annu Rev Ecol Syst. 1984;15:165–189.
- Ashraf SI, Kunes S. A trace of silence: memory and microRNA at the synapse. Curr Opin Neurobiol. 2006;16:535–539. - PubMed
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. - PubMed
- Baker M. MicroRNA profiling: separating signal from noise. Nat Methods. 2010;7:687–692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM086512/GM/NIGMS NIH HHS/United States
- 5RC1AR058681/AR/NIAMS NIH HHS/United States
- T32 GM008550/GM/NIGMS NIH HHS/United States
- R03 DC010244-03/DC/NIDCD NIH HHS/United States
- 5T32 GM 08550-15/GM/NIGMS NIH HHS/United States
- 1R01DK082605-01A1/DK/NIDDK NIH HHS/United States
- R03 DC010244-02/DC/NIDCD NIH HHS/United States
- R01 DK082605/DK/NIDDK NIH HHS/United States
- R03 DC010244-01/DC/NIDCD NIH HHS/United States
- R03 DC010244/DC/NIDCD NIH HHS/United States
- R03DC010244/DC/NIDCD NIH HHS/United States
- RC1 AR058681/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources