The human microbiome: at the interface of health and disease - PubMed (original) (raw)
Review
The human microbiome: at the interface of health and disease
Ilseung Cho et al. Nat Rev Genet. 2012.
Abstract
Interest in the role of the microbiome in human health has burgeoned over the past decade with the advent of new technologies for interrogating complex microbial communities. The large-scale dynamics of the microbiome can be described by many of the tools and observations used in the study of population ecology. Deciphering the metagenome and its aggregate genetic information can also be used to understand the functional properties of the microbial community. Both the microbiome and metagenome probably have important functions in health and disease; their exploration is a frontier in human genetics.
Figures
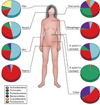
Figure 1. Compositional differences in the microbiome by anatomic site
High-throughput sequencing has revealed substantial intra-individual microbiome variation at different anatomical sites, and inter-individually for the same anatomical sites ,,,,,. However, higher level (e.g. phylum) taxonomic features display temporal (longitudinal) stability in individuals at specific anatomical sites. Such site-specific differences as well as observed conservation between human hosts provide an important framework to determine the biological and pathological significance of a particular microbiome composition. The figure indicates percentages of sequences at the taxonomic phylum level from selected references. Certain features, such as the presence or absence of Helicobacter pylori, can lead to permanent and marked perturbations in community composition.
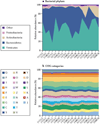
Figure 2. Conservation of bacterial genes despite taxonomic variation
A) Turnbaugh et al. studied the distal gut microbiome in lean and obese twins and their mothers. There were substantial and significant taxonomic variations amongst the individuals, although Firmicutes and Bacteroidetes still constituted the majority of the taxa. B) Through metagenomic analyses, the functional characteristics of the microbiomeas identified by COG pathways are largely conserved, despite the taxonomic variation. COG pathways are denoted by: S – Unknown; R – General function; L – DNA; G – Carbohydrates; E – Amino acids; M – Envelope; K – Transcription; J – Translation; C – Energy; T – Signal transduction; P – Inorganic; V – Defense; H – Coenzymes; O – Protein turnover; F – Nucleotides; U – Secretion; I – Lipids; D – Cell cycle; B – Chromatin; Q – Second metabolites; N – Cell motility; W – Extracellular; Z – Cytoskeleton; A – RNA. Reproduced with permission from Turnbaugh et al, Nature and the authors © Macmillan Publishers Ltd
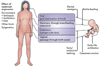
Figure 3. Acquisition of the microbiome in early life by vertical transmission and factors modifying mother-to-child microbial transmission
Through live-birth, mammals have important opportunities for mother → child microbial transmission, via direct-surface contact. However, many modern practices can reduce the organism and gene flow; several examples are illustrated. After initial introductions, there is strong selection by hosts for microbes with specific phenotypes, consistent with the extensive conservation shown in Figure 1. Acquisition is modified by offspring genetic and epigenetic differences (with respect to both maternal and paternal genes) that inform the competition for host resources by the vertically transmitted and environmentally acquired microbes. Ancestral organisms that have particular tissue- and niche-specific adaptations facilitate tissue tropisms and are selected, explaining the conserved niche-specificity compositions.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.
Ryan R, Hill S. Ryan R, et al. Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article. - Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Unlocking data: Decision-maker perspectives on cross-sectoral data sharing and linkage as part of a whole-systems approach to public health policy and practice.
Tweed E, Cimova K, Craig P, Allik M, Brown D, Campbell M, Henderson D, Mayor C, Meier P, Watson N. Tweed E, et al. Public Health Res (Southampt). 2024 Nov 20:1-30. doi: 10.3310/KYTW2173. Online ahead of print. Public Health Res (Southampt). 2024. PMID: 39582242 - Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.
Triana L, Palacios Huatuco RM, Campilgio G, Liscano E. Triana L, et al. Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
- Cangma Huadu granules attenuate H1N1 virus-induced severe lung injury correlated with repressed apoptosis and altered gut microbiome.
Liu M, Liu T, Wang X, Yu C, Qin T, Li J, Zhang M, Li Z, Cui X, Xu X, Liu Q. Liu M, et al. Front Microbiol. 2022 Aug 8;13:947112. doi: 10.3389/fmicb.2022.947112. eCollection 2022. Front Microbiol. 2022. PMID: 36090063 Free PMC article. - Gut microbiome composition differences among breeds impact feed efficiency in swine.
Bergamaschi M, Tiezzi F, Howard J, Huang YJ, Gray KA, Schillebeeckx C, McNulty NP, Maltecca C. Bergamaschi M, et al. Microbiome. 2020 Jul 22;8(1):110. doi: 10.1186/s40168-020-00888-9. Microbiome. 2020. PMID: 32698902 Free PMC article. - Metagenomic analysis reveals distinct patterns of gut lactobacillus prevalence, abundance, and geographical variation in health and disease.
Ghosh TS, Arnoux J, O'Toole PW. Ghosh TS, et al. Gut Microbes. 2020 Nov 9;12(1):1-19. doi: 10.1080/19490976.2020.1822729. Gut Microbes. 2020. PMID: 32985923 Free PMC article. - Microbial control of host gene regulation and the evolution of host-microbiome interactions in primates.
Grieneisen L, Muehlbauer AL, Blekhman R. Grieneisen L, et al. Philos Trans R Soc Lond B Biol Sci. 2020 Sep 28;375(1808):20190598. doi: 10.1098/rstb.2019.0598. Epub 2020 Aug 10. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 32772669 Free PMC article. Review. - Semi-supervised meta-learning elucidates understudied molecular interactions.
Wu Y, Xie L, Liu Y, Xie L. Wu Y, et al. Commun Biol. 2024 Sep 9;7(1):1104. doi: 10.1038/s42003-024-06797-z. Commun Biol. 2024. PMID: 39251833 Free PMC article.
References
- Baumann P, Moran NA. Non-cultivable microorganisms from symbiotic associations of insects and other hosts. Antonie van Leeuwenhoek. 1997;72:39–48. - PubMed
- Ehrlich SD. MetaHIT : The European Union Project on Metagenomics of the Human Intestinal Tract. In: Nelson KE, editor. Metagenomics of the Human Body. Springer; 2011. pp. 307–316.
- Ravel J, et al. Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 Suppl 1:4680–4687. Describes vaginal microbiome differences and similarities in women of reproductive age who vary by ethnicity, and explores factors related to bacterial vaginosis
- Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. Proposes enterotype classifications that are defined by the intrinsic characteristics of the gut microbiome, and that seem to be independent of ethnic or dietary factors
Publication types
MeSH terms
Grants and funding
- 5 P30 CA016087/CA/NCI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- 1UL1RR029893/RR/NCRR NIH HHS/United States
- R01 GM063270/GM/NIGMS NIH HHS/United States
- R01DK090989/DK/NIDDK NIH HHS/United States
- R01GM63270/GM/NIGMS NIH HHS/United States
- R01 DK090989/DK/NIDDK NIH HHS/United States
- UL1 RR029893/RR/NCRR NIH HHS/United States
- UH2 AR057506/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous