Major taste loss in carnivorous mammals - PubMed (original) (raw)
Major taste loss in carnivorous mammals
Peihua Jiang et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Mammalian sweet taste is primarily mediated by the type 1 taste receptor Tas1r2/Tas1r3, whereas Tas1r1/Tas1r3 act as the principal umami taste receptor. Bitter taste is mediated by a different group of G protein-coupled receptors, the Tas2rs, numbering 3 to ∼66, depending on the species. We showed previously that the behavioral indifference of cats toward sweet-tasting compounds can be explained by the pseudogenization of the Tas1r2 gene, which encodes the Tas1r2 receptor. To examine the generality of this finding, we sequenced the entire coding region of Tas1r2 from 12 species in the order Carnivora. Seven of these nonfeline species, all of which are exclusive meat eaters, also have independently pseudogenized Tas1r2 caused by ORF-disrupting mutations. Fittingly, the purifying selection pressure is markedly relaxed in these species with a pseudogenized Tas1r2. In behavioral tests, the Asian otter (defective Tas1r2) showed no preference for sweet compounds, but the spectacled bear (intact Tas1r2) did. In addition to the inactivation of Tas1r2, we found that sea lion Tas1r1 and Tas1r3 are also pseudogenized, consistent with their unique feeding behavior, which entails swallowing food whole without chewing. The extensive loss of Tas1r receptor function is not restricted to the sea lion: the bottlenose dolphin, which evolved independently from the sea lion but displays similar feeding behavior, also has all three Tas1rs inactivated, and may also lack functional bitter receptors. These data provide strong support for the view that loss of taste receptor function in mammals is widespread and directly related to feeding specializations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
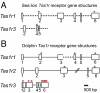
Widespread pseudogenization of the sweet-taste receptor gene Tas1r2 in 7 species within Carnivora. (A) Schematic diagram shows the positions of ORF-disrupting mutations in Tas1r2 from selected species within Carnivora. The intact dog Tas1r2 gene structure is shown as a reference. The positions where ORF-disrupting mutations occurred are marked with a red asterisk (*). (B) A 42-bp–long nucleotide sequence containing the ORF-disrupting mutation that occurs closest to 5′ end of the gene is shown for each species. The aligned dog sequence is shown above it, and the amino acid sequence deduced from the nucleotide sequence up to the mutation site is shown underneath it. The codon that contains the ORF-disrupting mutation (marked in red and underlined) is indicated by a box.
Fig. 2.
An evolutionary tree of Tas1r2 gene from 18 species within Carnivora. The evolutionary history is inferred by using the maximum-likelihood method based on the Tamura–Nei model (37) implemented in MEGA5 (26). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (2,000 replicates) is shown next to the branches (38). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Species with a pseudogenized Tas1r2 are marked with a diamond (red and gray depict species characterized in this study or previously, respectively). The human Tas1r2 is used as the outgroup for the analysis.
Fig. 3.
Sweet-taste preferences of two genotyped species. Two Asian otter and four spectacled bears were tested behaviorally for their preferences for sweeteners using a two-bowl preference setup. One bowl contained sweetener solution and the other contained plain water. Dashed line indicates no preference (50%). Sweeteners were tested at the following concentrations: fructose (0.8 M), galactose (0.8 M), lactose (0.5 M), maltose (0.7 M), sucrose (0.5 M), acesulfame-K (6.0 mM), aspartame (10 mM), neotame (10.5 mM), saccharin (6.2 mM), and sucralose (5.0 mM).
Fig. 4.
The sea lion and dolphin Tas1r receptor genes are inactivated by pseudogenization. (A) Sea lion Tas1r1 (Upper) and Tas1r3 (Lower) gene structures with boxes representing exons, and lines representing introns. Regions where ORF-disrupting mutations were found are marked with an asterisk. Regions with no sequence coverage are labeled with dashed lines. Only three exons have been sequenced for the sea lion Tas1r3. (B) Schematic drawings of the dolphin Tas1r1 (Top), Tas1r2 (Middle), and Tas1r3 (Bottom) gene structures. The symbols are the same as in A. The straight line with slashes indicates the intron is not proportionally scaled.
Fig. 5.
Dolphin _Tas2r_s are pseudogenes. (A) Schematic diagram shows the premature stop codons (either nonsense mutations or stop codons resulting from prior frameshift mutations, depicted with a red line) and only the frame-shift mutations (X) before the premature stop codons in dolphin _Tas2r_s. Tas2r receptor genes are shown in scale. No start codon is detected for Tas2r62bp. (B) A 42-bp–long nucleotide sequence containing the ORF-disrupting mutation that occurs closest to 5′ end of the gene is shown for each of the 10 identified dolphin Tas2r receptor genes; the aligned dog sequence is shown above it, and the amino acid sequence deduced from the nucleotide sequence up to the mutation site is shown underneath it. The codon that contains the ORF-disrupting mutation (marked in red and underlined) is indicated by a box.
Comment in
- Mismatches between feeding ecology and taste receptor evolution: an inconvenient truth.
Zhao H, Zhang J. Zhao H, et al. Proc Natl Acad Sci U S A. 2012 Jun 5;109(23):E1464; author reply E1465. doi: 10.1073/pnas.1205205109. Epub 2012 May 4. Proc Natl Acad Sci U S A. 2012. PMID: 22562799 Free PMC article. No abstract available.
Similar articles
- Evolution of the sweet taste receptor gene Tas1r2 in bats.
Zhao H, Zhou Y, Pinto CM, Charles-Dominique P, Galindo-González J, Zhang S, Zhang J. Zhao H, et al. Mol Biol Evol. 2010 Nov;27(11):2642-50. doi: 10.1093/molbev/msq152. Epub 2010 Jun 17. Mol Biol Evol. 2010. PMID: 20558596 Free PMC article. - Analyses of sweet receptor gene (Tas1r2) and preference for sweet stimuli in species of Carnivora.
Li X, Glaser D, Li W, Johnson WE, O'Brien SJ, Beauchamp GK, Brand JG. Li X, et al. J Hered. 2009 Jul-Aug;100 Suppl 1(Suppl 1):S90-100. doi: 10.1093/jhered/esp015. Epub 2009 Apr 14. J Hered. 2009. PMID: 19366814 Free PMC article. - Genomic and genetic evidence for the loss of umami taste in bats.
Zhao H, Xu D, Zhang S, Zhang J. Zhao H, et al. Genome Biol Evol. 2012;4(1):73-9. doi: 10.1093/gbe/evr126. Epub 2011 Nov 24. Genome Biol Evol. 2012. PMID: 22117084 Free PMC article. - Structure, function, and signaling of taste G-protein-coupled receptors.
Sanematsu K, Yoshida R, Shigemura N, Ninomiya Y. Sanematsu K, et al. Curr Pharm Biotechnol. 2014;15(10):951-61. doi: 10.2174/1389201015666140922105911. Curr Pharm Biotechnol. 2014. PMID: 25248559 Review. - Elucidation of mammalian bitter taste.
Meyerhof W. Meyerhof W. Rev Physiol Biochem Pharmacol. 2005;154:37-72. doi: 10.1007/s10254-005-0041-0. Rev Physiol Biochem Pharmacol. 2005. PMID: 16032395 Review.
Cited by
- Metabarcoding analysis provides insight into the link between prey and plant intake in a large alpine cat carnivore, the snow leopard.
Yoshimura H, Hayakawa T, Kikuchi DM, Zhumabai Uulu K, Qi H, Sugimoto T, Sharma K, Kinoshita K. Yoshimura H, et al. R Soc Open Sci. 2024 May 29;11(5):240132. doi: 10.1098/rsos.240132. eCollection 2024 May. R Soc Open Sci. 2024. PMID: 39076800 Free PMC article. - Membrane-bound chemoreception of bitter bile acids and peptides is mediated by the same subset of bitter taste receptors.
Schaefer S, Ziegler F, Lang T, Steuer A, Di Pizio A, Behrens M. Schaefer S, et al. Cell Mol Life Sci. 2024 May 15;81(1):217. doi: 10.1007/s00018-024-05202-6. Cell Mol Life Sci. 2024. PMID: 38748186 Free PMC article. - Synchronized Expansion and Contraction of Olfactory, Vomeronasal, and Taste Receptor Gene Families in Hystricomorph Rodents.
Niimura Y, Biswa BB, Kishida T, Toyoda A, Fujiwara K, Ito M, Touhara K, Inoue-Murayama M, Jenkins SH, Adenyo C, Kayang BB, Koide T. Niimura Y, et al. Mol Biol Evol. 2024 Apr 2;41(4):msae071. doi: 10.1093/molbev/msae071. Mol Biol Evol. 2024. PMID: 38649162 Free PMC article. - Insights to Study, Understand and Manage Extruded Dry Pet Food Palatability.
Le Guillas G, Vanacker P, Salles C, Labouré H. Le Guillas G, et al. Animals (Basel). 2024 Apr 3;14(7):1095. doi: 10.3390/ani14071095. Animals (Basel). 2024. PMID: 38612333 Free PMC article. Review. - Diversity and evolution of the vertebrate chemoreceptor gene repertoire.
Policarpo M, Baldwin MW, Casane D, Salzburger W. Policarpo M, et al. Nat Commun. 2024 Feb 15;15(1):1421. doi: 10.1038/s41467-024-45500-y. Nat Commun. 2024. PMID: 38360851 Free PMC article.
References
- Max M, et al. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. - PubMed
- Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat Neurosci. 2001;4:492–498. - PubMed
- Nelson G, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK094759/DK/NIDDK NIH HHS/United States
- R01 DC003055/DC/NIDCD NIH HHS/United States
- R01 DC004698/DC/NIDCD NIH HHS/United States
- R01 DK058797/DK/NIDDK NIH HHS/United States
- R01 DC003155/DC/NIDCD NIH HHS/United States
- R01 DC010842/DC/NIDCD NIH HHS/United States
- DC010842/DC/NIDCD NIH HHS/United States
- R03 DC003509/DC/NIDCD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous