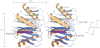Structure of a yeast Dyn2-Nup159 complex and molecular basis for dynein light chain-nuclear pore interaction - PubMed (original) (raw)
Structure of a yeast Dyn2-Nup159 complex and molecular basis for dynein light chain-nuclear pore interaction
Erin M Romes et al. J Biol Chem. 2012.
Abstract
The nuclear pore complex gates nucleocytoplasmic transport through a massive, eight-fold symmetric channel capped by a nucleoplasmic basket and structurally unique, cytoplasmic fibrils whose tentacles bind and regulate asymmetric traffic. The conserved Nup82 complex, composed of Nsp1, Nup82, and Nup159, forms the unique cytoplasmic fibrils that regulate mRNA nuclear export. Although the nuclear pore complex plays a fundamental, conserved role in nuclear trafficking, structural information about the cytoplasmic fibrils is limited. Here, we investigate the structural and biochemical interactions between Saccharomyces cerevisiae Nup159 and the nucleoporin, Dyn2. We find that Dyn2 is predominantly a homodimer and binds arrayed sites on Nup159, promoting the Nup159 parallel homodimerization. We present the first structure of Dyn2, determined at 1.85 Å resolution, complexed with a Nup159 target peptide. Dyn2 resembles homologous metazoan dynein light chains, forming homodimeric composite substrate binding sites that engage two independent 10-residue target motifs, imparting a β-strand structure to each peptide via antiparallel extension of the Dyn2 core β-sandwich. Dyn2 recognizes a highly conserved QT motif while allowing sequence plasticity in the flanking residues of the peptide. Isothermal titration calorimetric analysis of the comparative binding of Dyn2 to two Nup159 target sites shows similar affinities (18 and 13 μM), but divergent thermal binding modes. Dyn2 homodimers are arrayed in the crystal lattice, likely mimicking the arrayed architecture of Dyn2 on the Nup159 multivalent binding sites. Crystallographic interdimer interactions potentially reflect a cooperative basis for Dyn2-Nup159 complex formation. Our data highlight the determinants that mediate oligomerization of the Nup82 complex and promote a directed, elongated cytoplasmic fibril architecture.
Figures
FIGURE 1.
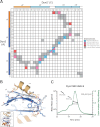
S. cerevisiae Dyn2 is a conserved dynein light chain involved in diverse macromolecular complexes including the nuclear pore complex and cytoplasmic dynein motor complex. A, sequence alignment of 12 dynein light chain family members ranging from S. cerevisiae to human. Residues aligned with 100 and 80% identity are colored green and yellow, respectively. Amino acid numbers and secondary structure elements, based on the S. cerevisiae Dyn2 structure, are shown above the alignment. Residues involved in Dyn2 dimerization and Dyn2-Nup159 pep2 binding are indicated below the alignment by asterisks based on European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) PDBe Protein Interfaces, Surface and Assemblies (PISA). Black, Dyn2-Dyn2 chain A-C interactions; red, Dyn2-Nup159_2 chain A-D or chain C-B interactions; blue, Dyn2-Nup159_2 chain A-B or chain C-D interactions. Solvent-accessible (SA) surface area for respective Dyn2 chain C residues is indicated below the alignment, calculated in the presence (black) and absence of the Nup159_2 chain D (gray) using the Accessible Surface Area Analysis tool in CCP4 (41). A. gossypii, Ashbya gossypii; C. dubliniensis, Candida dubliniensis; S. pombe, Schizosaccharomyces pombe; D. discoideum, Dictyostelium discoideum; D. rerio, Danio rerio; X. laevis, Xenopus laevis; R. norvegicus, Rattus norvegicus; P. troglodytes, Pan troglodytes; H. sapiens, Homo sapiens. B, diagram of the nuclear pore complex illustrating the cytoplasmic localization of Nup159 and the Nup82 complex to cytoplasmic fibrils. C, domain architecture of known Dyn2-binding proteins: Nup159 and Pac11. Nup159 is composed of an N-terminal (N-term) β-propeller domain involved in Dbp5 binding, central Phe-Gly-rich repeats common to nucleoporins, a DID composed of five QT consensus motifs (residues 1103–1177) with Dyn2 binding activity, and a C-terminal (C-term) region involved in Nup159 anchoring to the nuclear pore complex (11, 14, 15). Pac11, the yeast dynein intermediate chain, shares architectural similarities with Nup159, composed of an N-terminal coiled-coil domain, tandem Dyn2 QT binding motifs, and a C-terminal WD-40 repeat domain, predicted to be a β-propeller. D, sequence alignment of the Dyn2 binding motifs from Nup159 and Pac11 highlighting the invariant QT motif. Nup159_2 secondary structure is indicated above the alignment. Nup159_2 residues involved in Dyn2 binding are indicated by asterisks below the alignment, as is solvent-accessible surface area, calculated in the presence (black) and absence of Dyn2 chains (gray).
FIGURE 2.
Structure of Dyn2-Nup159 pep2 complex shows a quaternary complex composed of a Dyn2 dimer, bound to two Nup159 peptides through parallel, composite β-sheets. A, diagram of the Dyn2-Nup159 complex. Dyn2 chain A is shown in orange (α-helices) and dark blue (β-strands), and Dyn2 chain C is shown in beige (α-helices) and light blue (β-strands). The two-fold noncrystallographic symmetry operator that relates the Dyn2 and Nup159 chains in the asymmetric unit is indicated about the z axis. The image at right shows the complex after a 90° rotation about the y axis. B, the complex as shown in the two orientations in A, with Dyn2 chain A superimposed on the human dynein light chain, LC8 (light green) bound to a PIN peptide (yellow) (PDB 1CMI) after a least squares fit with an r.m.s.d. of 0.6 Å over 87 aligned Cα atoms (34). Helices are shown in cylindrical format. Structural differences between Dyn2 and human LC8 are indicated by red arrows and are dominated by loop regions as well as the bound peptides.
FIGURE 3.
Dyn2 homodimerizes via an extensive network of van der Waals contacts and hydrogen bonds. A, interaction matrix, showing the pseudo-symmetric bonding and contact networks formed between Dyn2 protomers A and C in the complex. Secondary structure elements corresponding to the residues of each protomer are indicated along the axes of the matrix. Backbone-backbone, backbone-side chain, side chain-side chain, and van der Waals interactions are indicated in blue, pink, red, and gray, respectively, and correlate with distances less than or equal to 3.5 (hydrogen bonds) and 4.5 Å (van der Waals contacts). Numbers in cells indicate the total number of hydrogen bonds (greater than one) between 2 residues. B, diagram of key residues and structural elements involved in the Dyn2-Dyn2 interface. The Dyn2 homodimer is shown as colored in Fig. 1. Specific Dyn2 residues mediating homodimerization are shown in stick format. Hydrogen bonds are indicated as dashed lines. The interface involves extensive antiparallel β-strand-β-strand interactions as well as contributions from the α2 helices that flank the central β-sandwich. The inset shows the relative orientation of the complex. C, size exclusion chromatography and multiangle light scattering (SEC-MALS) analysis of Dyn2, injected at an initial concentration of 200 μ
m
(green) in 100 μl. The Raleigh Ratio elution profile was normalized. Dyn2 predominantly forms a dimer in solution at pH 6.8, with additional, higher order tetrameric and octameric species detected as well. The Dyn2 construct analyzed has a calculated monomeric molecular mass of 10,852 Da.
FIGURE 4.
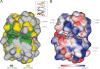
Dyn2 binds substrates through a highly conserved, positively charged composite groove formed by Dyn2 dimerization. A, conservation, as highlighted in Fig. 1_A_ (green, 100% identity; yellow, 80% identity, as determined across 12 diverse species), mapped on the Dyn2 dimer shown in surface representation. Nup159 pep2 is shown in stick format in purple, inserted in the highly conserved interdimer groove. The highly conserved QT substrate motif (green sticks) is located C-terminal to the Nup159_2 β-strand. Conservation, however, is equally distributed across the Dyn2 substrate binding region. The inset shows the relative orientation of the complex in graphic format colored as in Fig. 2_A. B_, electrostatic surface calculated using APBS to generate solvent-accessible surface potentials that are shown in kBT/e, colored according to the key shown (42). Nup159 pep2 is shown in purple stick format with specific Dyn2 residues involved in hydrogen bond contacts labeled. The conserved QT motif is shown in green stick format. Dyn2-Nup159 pep2 interactions include Tyr-68–Glu-1119, Glu-38′–Gln-1123, and Lys-12–Asp-1125. The complex is oriented as in A.
FIGURE 5.
The Dyn2-Nup159 pep2 interaction is mediated by an extensive interaction network that recognizes 10 contiguous Nup159 residues, dually conferring specificity and substrate plasticity. A and B, close-up of residues involved in the Dyn2-Nup159 pep2 interaction. Secondary structure elements are shown as in Fig. 2_A_, with specific residues that mediate the Dyn2-Nup159 pep2 interaction shown in stick format and their corresponding hydrogen-bonding network shown with dashed lines. C, interaction matrix, showing the contact networks formed between Dyn2 protomers A and C with Nup159. Secondary structure elements and protomer designation are indicated along matrix axes. Backbone-backbone, backbone-side chain, side chain-side chain, and van der Waals interactions are indicated in blue, pink, red, and gray, respectively, and correlate with distances less than or equal to 3.5 (hydrogen bonds) and 4.5 Å (van der Waals contacts).
FIGURE 6.
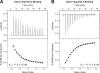
The Dyn2 interactions with Nup159 pep2 and pep4 occur in a 1:1 stoichiometry and exhibit similar affinities but differ in their thermal binding modes. A, 17 × 2 μl of 1 m
m
Nup159 pep2 was injected into 200 μl of 100 μ
m
Dyn2. The thermogram (upper panel) displays μcal/sec over the injection period (min). B, 17 × 2 μl of 1 m
m
Nup159 pep4 was injected into 200 μl of 100 μ
m
Dyn2. Dyn2 binding to Nup159 pep2 (A) displayed an endothermic binding isotherm, whereas Dyn2 binding to Nup159 pep4 (B) showed exothermic binding. Thermograms (upper panels) were integrated, and the resulting isotherm was fit to a one-site binding model (lower panels) through iterative fitting. KD values presented (inset, lower panel) are the average of three independent experiments: Dyn2-Nup159 pep2, KD = 17.9 ± 3.8 μ
m
, Δ_H_ = 2500 cal/mol, Δ_S_ = 31 cal/mol/deg, n = 0.33 sites; Dyn2-Nup159 pep4, KD = 13.1 ± 1.6 μ
m
, Δ_H_ = −4000 cal/mol, Δ_S_ = 8.6 cal/mol/deg, n = 0.39 sites.
FIGURE 7.
Crystallographic contacts array Dyn2 dimers linearly in an arrangement that affords polarized binding to arrayed Dyn2 binding motifs. Dyn2-Nup159 crystallographic symmetry mates are shown in graphic representation, colored as in Fig. 2_A_. Dyn2 interdimer interactions coupled with parallel, arrayed binding motifs on Nup159 likely promote linear, cooperative binding activity between Dyn2 dimers and Nup159.
Similar articles
- Multiple recognition motifs in nucleoporin Nup159 provide a stable and rigid Nup159-Dyn2 assembly.
Nyarko A, Song Y, Nováček J, Žídek L, Barbar E. Nyarko A, et al. J Biol Chem. 2013 Jan 25;288(4):2614-22. doi: 10.1074/jbc.M112.432831. Epub 2012 Dec 8. J Biol Chem. 2013. PMID: 23223634 Free PMC article. - Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex.
Stelter P, Kunze R, Flemming D, Höpfner D, Diepholz M, Philippsen P, Böttcher B, Hurt E. Stelter P, et al. Nat Cell Biol. 2007 Jul;9(7):788-96. doi: 10.1038/ncb1604. Epub 2007 Jun 3. Nat Cell Biol. 2007. PMID: 17546040 - Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex.
Yoshida K, Seo HS, Debler EW, Blobel G, Hoelz A. Yoshida K, et al. Proc Natl Acad Sci U S A. 2011 Oct 4;108(40):16571-6. doi: 10.1073/pnas.1112846108. Epub 2011 Sep 19. Proc Natl Acad Sci U S A. 2011. PMID: 21930948 Free PMC article. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review.
Cited by
- Multivalent IDP assemblies: Unique properties of LC8-associated, IDP duplex scaffolds.
Clark SA, Jespersen N, Woodward C, Barbar E. Clark SA, et al. FEBS Lett. 2015 Sep 14;589(19 Pt A):2543-51. doi: 10.1016/j.febslet.2015.07.032. Epub 2015 Jul 29. FEBS Lett. 2015. PMID: 26226419 Free PMC article. Review. - Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform.
Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, Williams R, Rice WJ, Stokes DL, Zenklusen D, Chait BT, Sali A, Rout MP. Fernandez-Martinez J, et al. Cell. 2016 Nov 17;167(5):1215-1228.e25. doi: 10.1016/j.cell.2016.10.028. Epub 2016 Nov 10. Cell. 2016. PMID: 27839866 Free PMC article. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review. - The Structure Inventory of the Nuclear Pore Complex.
Schwartz TU. Schwartz TU. J Mol Biol. 2016 May 22;428(10 Pt A):1986-2000. doi: 10.1016/j.jmb.2016.03.015. Epub 2016 Mar 22. J Mol Biol. 2016. PMID: 27016207 Free PMC article. Review. - Characterization of a novel interaction of the Nup159 nucleoporin with asymmetrically localized spindle pole body proteins and its link with autophagy.
de Oya IG, Manzano-López J, Álvarez-Llamas A, Vázquez-Aroca MP, Cepeda-García C, Monje-Casas F. de Oya IG, et al. PLoS Biol. 2023 Aug 3;21(8):e3002224. doi: 10.1371/journal.pbio.3002224. eCollection 2023 Aug. PLoS Biol. 2023. PMID: 37535687 Free PMC article.
References
- Schwartz T. U. (2005) Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 15, 221–226 - PubMed
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 - PubMed
- Gerace L. (1988) Functional organization of the nuclear envelope. Ann. Rev. Cell Biol. 4, 335–374 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases