A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells - PubMed (original) (raw)
A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells
Kyle J Roux et al. J Cell Biol. 2012.
Abstract
We have developed a new technique for proximity-dependent labeling of proteins in eukaryotic cells. Named BioID for proximity-dependent biotin identification, this approach is based on fusion of a promiscuous Escherichia coli biotin protein ligase to a targeting protein. BioID features proximity-dependent biotinylation of proteins that are near-neighbors of the fusion protein. Biotinylated proteins may be isolated by affinity capture and identified by mass spectrometry. We apply BioID to lamin-A (LaA), a well-characterized intermediate filament protein that is a constituent of the nuclear lamina, an important structural element of the nuclear envelope (NE). We identify multiple proteins that associate with and/or are proximate to LaA in vivo. The most abundant of these include known interactors of LaA that are localized to the NE, as well as a new NE-associated protein named SLAP75. Our results suggest BioID is a useful and generally applicable method to screen for both interacting and neighboring proteins in their native cellular environment.
Figures
Figure 1.
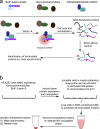
Model for application of BioID method. (a) Expression of a promiscuous biotin–ligase fusion protein in live cells leads to the selective biotinylation of proteins proximate to that fusion protein. After stringent cell lysis and protein denaturation, biotinylated proteins are affinity purified. These candidate proteins can be identified by mass spectrometry or immunoblot analysis. (b) In our application of BioID to LaA to identify candidate proteins we used HEK293 cells stably expressing inducible mycBirA*LaA. 24 h before lysis, cells were induced to express mycBirA*LaA with doxycycline and to biotinylate endogenous proteins with 50 µM biotin. Cells were lysed under stringent conditions and biotinylated proteins collected on streptavidin-conjugated beads for subsequent analysis and identification.
Figure 2.
BirA* promiscuously biotinylates endogenous proteins in mammalian cells. HeLa cells were analyzed 24 h after transient transfection with myc-BirA-WT or myc-BirA* (R118G). After transfection, cells were cultured either with or without supplemental biotin (50 µM). (a) By Western blot analysis similar levels of the exogenous BirA (asterisk) are detected in all samples with anti-myc. Biotinylated proteins, including both exogenous BirA (asterisk) and endogenous proteins, were detected with HRP-streptavidin. Enhanced protein biotinylation is observed in the myc-BirA* samples as compared with the WT isoform. This difference is dramatically enhanced by the presence of excess biotin. (b) By fluorescence microscopy the BirA is predominantly nuclear as observed with anti-myc (red). Biotinylated proteins were detected with fluorescently labeled streptavidin (green). Considerable biotinylation is only observed in cells expressing myc-BirA* and supplemented with excess biotin. The biotin signal predominantly colocalizes with myc-BirA*. DNA is labeled with Hoechst (blue). Bar, 4 µm.
Figure 3.
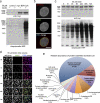
Proximity-dependent promiscuous biotinylation by BioID-LaA. HEK293 cells inducibly expressing myc-BirA*LaA, or parental controls, were analyzed 24 h after induction with or without excess biotin. (a) By immunoblot analysis the LaA fusion protein (asterisk) is detected with anti-myc. In control cells levels of endogenously biotinylated proteins are unaffected by the supplemental biotin; however, the biotinylation of endogenous proteins by myc-BirA*LaA is dramatically enhanced in the presence of excess biotin. (b) The myc-BirA*LaA (red) is detected at the nuclear rim and to a lesser extent in the nucleoplasm. Biotinylated proteins (green) colocalize with the LaA fusion protein. DNA is labeled with Hoechst (blue). Bar, 5 µm. (c) To monitor the relative rate of biotinylation by BioID, myc-BirA*LaA HEK293 cells were provided with 50 µM biotin at different time points. The levels of myc-BirA*LaA (asterisk in panel a) are similar for all conditions; however, the extent of biotinylation increases with duration of biotin supplementation until it is saturated by 24 h. (d) Similar results were observed by fluorescence microscopy. Myc-BirA*LaA was detected with anti-myc (red) and biotinylated proteins with fluorescently labeled streptavidin (green). DNA was labeled with Hoechst (blue). Bar, 25 µm. (e) The identity of the proteins biotinylated by BioID-LaA was determined by mass spectrometry of proteins isolated with streptavidin-coupled magnetic beads. The relative abundance (percentage of total identified spectra adjusted for protein size, identified spectra per 103 amino acids) and classification of proteins uniquely biotinylated by myc-BirA*LaA is depicted in the chart. Conservatively estimated, 50% of the detected proteins predominantly reside at the nuclear lamina, INM, or nucleoplasmic face of the NPCs.
Figure 4.
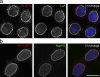
SLAP75 is a novel peripheral membrane constituent of the NE identified with BioID-LaA. To determine if FAM169A/SLAP75 is a novel peripheral membrane constituent of the NE its subcellular localization was analyzed by immunofluorescence microcopy. (a) As detected with anti-SLAP75 (red), the endogenous protein is colocalized with LaA (green) at the NE of HEK293 cells. (b) Although endogenous SLAP75 is not detected in HeLa cells (not depicted), transiently expressed HA-SLAP75 (red) detected with anti-FAM169A/SLAP75 is localized to the NE, labeled with anti-Nup153 (green). DNA was labeled with Hoechst (blue). Bar, 10 µm.
Similar articles
- An improved smaller biotin ligase for BioID proximity labeling.
Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. Kim DI, et al. Mol Biol Cell. 2016 Apr 15;27(8):1188-96. doi: 10.1091/mbc.E15-12-0844. Epub 2016 Feb 24. Mol Biol Cell. 2016. PMID: 26912792 Free PMC article. - BioID: A Screen for Protein-Protein Interactions.
Roux KJ, Kim DI, Burke B, May DG. Roux KJ, et al. Curr Protoc Protein Sci. 2018 Feb 21;91:19.23.1-19.23.15. doi: 10.1002/cpps.51. Curr Protoc Protein Sci. 2018. PMID: 29516480 Free PMC article. - Biotin Proximity Labeling for Protein-Protein Interaction Discovery: The BioID Method.
Habel JE. Habel JE. Methods Mol Biol. 2021;2261:357-379. doi: 10.1007/978-1-0716-1186-9_22. Methods Mol Biol. 2021. PMID: 33421001 - Biotin protein ligase as you like it: Either extraordinarily specific or promiscuous protein biotinylation.
Cronan JE. Cronan JE. Proteins. 2024 Apr;92(4):435-448. doi: 10.1002/prot.26642. Epub 2023 Nov 23. Proteins. 2024. PMID: 37997490 Review. - Proteomic navigation using proximity-labeling.
Gentzel M, Pardo M, Subramaniam S, Stewart AF, Choudhary JS. Gentzel M, et al. Methods. 2019 Jul 15;164-165:67-72. doi: 10.1016/j.ymeth.2019.03.028. Epub 2019 Apr 4. Methods. 2019. PMID: 30953756 Review.
Cited by
- Protein tyrosine phosphatases in cell adhesion.
Young KA, Biggins L, Sharpe HJ. Young KA, et al. Biochem J. 2021 Mar 12;478(5):1061-1083. doi: 10.1042/BCJ20200511. Biochem J. 2021. PMID: 33710332 Free PMC article. Review. - Spatial proximity of proteins surrounding zyxin under force-bearing conditions.
Cheah JS, Jacobs KA, Lai TW, Caballelo R, Yee JL, Ueda S, Heinrich V, Yamada S. Cheah JS, et al. Mol Biol Cell. 2021 Jun 15;32(13):1221-1228. doi: 10.1091/mbc.E19-10-0568. Epub 2021 Apr 28. Mol Biol Cell. 2021. PMID: 33909446 Free PMC article. - RAD51AP1 regulates ALT-HDR through chromatin-directed homeostasis of TERRA.
Kaminski N, Wondisford AR, Kwon Y, Lynskey ML, Bhargava R, Barroso-González J, García-Expósito L, He B, Xu M, Mellacheruvu D, Watkins SC, Modesti M, Miller KM, Nesvizhskii AI, Zhang H, Sung P, O'Sullivan RJ. Kaminski N, et al. Mol Cell. 2022 Nov 3;82(21):4001-4017.e7. doi: 10.1016/j.molcel.2022.09.025. Epub 2022 Oct 19. Mol Cell. 2022. PMID: 36265488 Free PMC article. - RNA-protein interaction mapping via MS2- or Cas13-based APEX targeting.
Han S, Zhao BS, Myers SA, Carr SA, He C, Ting AY. Han S, et al. Proc Natl Acad Sci U S A. 2020 Sep 8;117(36):22068-22079. doi: 10.1073/pnas.2006617117. Epub 2020 Aug 24. Proc Natl Acad Sci U S A. 2020. PMID: 32839320 Free PMC article. - Lamin A/C-dependent interaction with 53BP1 promotes cellular responses to DNA damage.
Gibbs-Seymour I, Markiewicz E, Bekker-Jensen S, Mailand N, Hutchison CJ. Gibbs-Seymour I, et al. Aging Cell. 2015 Apr;14(2):162-9. doi: 10.1111/acel.12258. Epub 2015 Jan 23. Aging Cell. 2015. PMID: 25645366 Free PMC article.
References
- Bodoor K., Shaikh S., Salina D., Raharjo W.H., Bastos R., Lohka M., Burke B. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials