Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation - PubMed (original) (raw)
Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation
Daniel M Passon et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Proteins of the Drosophila behavior/human splicing (DBHS) family include mammalian SFPQ (PSF), NONO (p54nrb), PSPC1, and invertebrate NONA and Hrp65. DBHS proteins are predominately nuclear, and are involved in transcriptional and posttranscriptional gene regulatory functions as well as DNA repair. DBHS proteins influence a wide gamut of biological processes, including the regulation of circadian rhythm, carcinogenesis, and progression of cancer. Additionally, mammalian DBHS proteins associate with the architectural long noncoding RNA NEAT1 (Menε/β) to form paraspeckles, subnuclear bodies that alter gene expression via the nuclear retention of RNA. Here we describe the crystal structure of the heterodimer of the multidomain conserved region of the DBHS proteins, PSPC1 and NONO. These proteins form an extensively intertwined dimer, consistent with the observation that the different DBHS proteins are typically copurified from mammalian cells, and suggesting that they act as obligate heterodimers. The PSPC1/NONO heterodimer has a right-handed antiparallel coiled-coil that positions two of four RNA recognition motif domains in an unprecedented arrangement on either side of a 20-Å channel. This configuration is supported by a protein:protein interaction involving the NONA/paraspeckle domain, which is characteristic of the DBHS family. By examining various mutants and truncations in cell culture, we find that DBHS proteins require an additional antiparallel coiled-coil emanating from either end of the dimer for paraspeckle subnuclear body formation. These results suggest that paraspeckles may potentially form through self-association of DBHS dimers into higher-order structures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
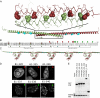
Fig. 1.
Structure of a PSPC1/NONO heterodimer. (A) Domain structure indicating RNA recognition motifs 1 and 2, NOPS domain, and coiled-coil. (B) Stereoview of PSPC1 (cartoon) and NONO (surface). (C) Side (Left) and top (Right) views of PSPC1 (red) and NONO (green). (D) Cutaway view revealing voids (putative RNA-binding residues, cyan). (E) Side view highlighting putative RNA-binding residues of RRM1 (cyan).
Fig. 2.
Conserved residues in the NOPS domain (NONO: Y267, W271; PSPC1: Y275, W279) are critical for dimerization and paraspeckle localization. (A) The NOPS domain of PSPC1 (yellow) with Y275 and W279 circled. (B) Conserved Tyr and Trp residues have radically different side-chain conformations in NONO and PSPC1. (C) Wild-type Tyr and Trp, and not Ala, are required for paraspeckle formation by full-length YFP-NONO and mCherry-PSPC1 fusion proteins in HeLa cells. (Scale bars: 5 μm.) (D) Wild-type Tyr and Trp, and not Ala, are necessary for hetero- and homodimer formation in yeast two-hybrid analyses of full-length PSPC1/NONO (EYFP as negative control).
Fig. 3.
Coiled-coil formation is critical for paraspeckle targeting by DBHS proteins. (A) Observed coiled-coil (atomic model) and model of extended coiled-coil (cyan residues indicate hendecad repeat). (B) Sequence alignment of PSPC1, NONO, and SFPQ coiled coils; green arrows mark C termini of truncated PSPC1 proteins able to form paraspeckles; red arrows, those that cannot. (C) A schematic model for coiled-coil-mediated oligomerization of DBHS proteins. (D) YFP fusion proteins of PSPC1 proteins terminating after residue 337 colocalize with paraspeckles in HeLa cells; shorter proteins do not. (Scale bars: 5 μm.) (E) Wild-type endogenous NONO is coimmunoprecipitated by truncated YFP-PSPC1 proteins in an anti-GFP pull-down. Anti-GFP and anti-NONO Western blots.
Similar articles
- Crystallization of a paraspeckle protein PSPC1-NONO heterodimer.
Passon DM, Lee M, Fox AH, Bond CS. Passon DM, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Oct 1;67(Pt 10):1231-4. doi: 10.1107/S1744309111026212. Epub 2011 Sep 29. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 22102035 Free PMC article. - A conserved charged single α-helix with a putative steric role in paraspeckle formation.
Dobson L, Nyitray L, Gáspári Z. Dobson L, et al. RNA. 2015 Dec;21(12):2023-9. doi: 10.1261/rna.053058.115. Epub 2015 Oct 1. RNA. 2015. PMID: 26428695 Free PMC article. - Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
Lee PW, Marshall AC, Knott GJ, Kobelke S, Martelotto L, Cho E, McMillan PJ, Lee M, Bond CS, Fox AH. Lee PW, et al. J Biol Chem. 2022 Nov;298(11):102563. doi: 10.1016/j.jbc.2022.102563. Epub 2022 Oct 7. J Biol Chem. 2022. PMID: 36209820 Free PMC article. - Paraspeckles: nuclear bodies built on long noncoding RNA.
Bond CS, Fox AH. Bond CS, et al. J Cell Biol. 2009 Sep 7;186(5):637-44. doi: 10.1083/jcb.200906113. Epub 2009 Aug 31. J Cell Biol. 2009. PMID: 19720872 Free PMC article. Review. - Involvement of paraspeckle components in viral infections.
Milcamps R, Michiels T. Milcamps R, et al. Nucleus. 2024 Dec;15(1):2350178. doi: 10.1080/19491034.2024.2350178. Epub 2024 May 8. Nucleus. 2024. PMID: 38717150 Free PMC article. Review.
Cited by
- Pre-mRNA processing factors meet the DNA damage response.
Montecucco A, Biamonti G. Montecucco A, et al. Front Genet. 2013 Jun 6;4:102. doi: 10.3389/fgene.2013.00102. eCollection 2013. Front Genet. 2013. PMID: 23761808 Free PMC article. - Swine NONO promotes IRF3-mediated antiviral immune response by Detecting PRRSV N protein.
Jiang D, Sui C, Wu X, Jiang P, Bai J, Hu Y, Cong X, Li J, Yoo D, Miller LC, Lee C, Du Y, Qi J. Jiang D, et al. PLoS Pathog. 2024 Oct 16;20(10):e1012622. doi: 10.1371/journal.ppat.1012622. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39413144 Free PMC article. - The tumor suppressor annexin A10 is a novel component of nuclear paraspeckles.
Quiskamp N, Poeter M, Raabe CA, Hohenester UM, König S, Gerke V, Rescher U. Quiskamp N, et al. Cell Mol Life Sci. 2014 Jan;71(2):311-29. doi: 10.1007/s00018-013-1375-4. Epub 2013 May 29. Cell Mol Life Sci. 2014. PMID: 23715859 Free PMC article. - Anti-cancer effect of low dose of celecoxib may be associated with lnc-SCD-1:13 and lnc-PTMS-1:3 but not COX-2 in NCI-N87 cells.
Song B, Shu ZB, Du J, Ren JC, Feng Y. Song B, et al. Oncol Lett. 2017 Aug;14(2):1775-1779. doi: 10.3892/ol.2017.6316. Epub 2017 Jun 6. Oncol Lett. 2017. PMID: 28789408 Free PMC article. - NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing.
Jiang L, Shao C, Wu QJ, Chen G, Zhou J, Yang B, Li H, Gou LT, Zhang Y, Wang Y, Yeo GW, Zhou Y, Fu XD. Jiang L, et al. Nat Struct Mol Biol. 2017 Oct;24(10):816-824. doi: 10.1038/nsmb.3455. Epub 2017 Aug 28. Nat Struct Mol Biol. 2017. PMID: 28846091 Free PMC article.
References
- Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. - PubMed
- Brown SA, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. - PubMed
- Stanewsky R, Rendahl KG, Dill M, Saumweber H. Genetic and molecular analysis of the X chromosomal region 14B17-14C4 in Drosophila melanogaster: Loss of function in NONA, a nuclear protein common to many cell types, results in specific physiological and behavioral defects. Genetics. 1993;135:419–442. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous