The prolactin gene: a paradigm of tissue-specific gene regulation with complex temporal transcription dynamics - PubMed (original) (raw)
Review
The prolactin gene: a paradigm of tissue-specific gene regulation with complex temporal transcription dynamics
K Featherstone et al. J Neuroendocrinol. 2012 Jul.
Free PMC article
Abstract
Transcription of numerous mammalian genes is highly pulsatile, with bursts of expression occurring with variable duration and frequency. The presence of this stochastic or 'noisy' expression pattern has been relatively unexplored in tissue systems. The prolactin gene provides a model of tissue-specific gene regulation resulting in pulsatile transcription dynamics in both cell lines and endocrine tissues. In most cell culture models, prolactin transcription appears to be highly variable between cells, with differences in transcription pulse duration and frequency. This apparently stochastic transcription is constrained by a transcriptional refractory period, which may be related to cycles of chromatin remodelling. We propose that prolactin transcription dynamics result from the summation of oscillatory cellular inputs and by regulation through chromatin remodelling cycles. Observations of transcription dynamics in cells within pituitary tissue show reduced transcriptional heterogeneity and can be grouped into a small number of distinct patterns. Thus, it appears that the tissue environment is able to reduce transcriptional noise to enable coordinated tissue responses to environmental change. We review the current knowledge on the complex tissue-specific regulation of the prolactin gene in pituitary and extra-pituitary sites, highlighting differences between humans and rodent experimental animal models. Within this context, we describe the transcription dynamics of prolactin gene expression and how this may relate to specific processes occurring within the cell.
© 2012 The Authors. Journal of Neuroendocrinology © 2012 Blackwell Publishing Ltd.
Figures
Fig. 1
The prolactin gene locus. (
a
) The location and organisation of the human, mouse and rat prolactin loci (not to scale). The loci are orientated by the prolactin gene and this is reflected by the numbering of the locus position on the chromosome. The diagram illustrates the vastly different structure of the prolactin loci between primate and rodent species. Data from ensembl (human: release GRCh37, rat: release RGSC3.4, mouse: release NCBIM37) and (6,7). HDGFL1, hepatoma-derived growth factor like 1. (
b
) Alignment of the repeatmasked human prolactin gene ± 10 kbp sequence to the rat prolactin gene ± 10 kbp sequence shows sequence homology in noncoding sequences in upstream regulatory regions and in gene introns. Alignment generated using Pipmaker (8). (
c
) Repeat composition of the human and rat prolactin loci. The rat prolactin locus shows a substantial difference in repeat content in comparison with the genome average. Data derived from repeatmasker [A. F. A. Smit, R. Hubley and P. Green, unpublished data. Version: open-3.3.0 (RMLib: 20110419)] using ABBlast and default settings (10,11).
Fig. 2
Schematic of regulatory elements in the human prolactin locus. (
a
) Organisation of regulatory elements in the prolactin locus. Exons (black boxes), promoters (white boxes), enhancers (green boxes) and silencers (red boxes) are shown. (
b
) Organisation of regulatory elements that facilitate expression in the pituitary. Transcription factors with demonstrated binding to the prolactin locus are listed. (
c
) Organisation of regulatory elements that facilitates expression in nonpituitary tissues via the upstream alternative promoter and exon1a. Elements with potentially specific activity within different extra-pituitary tissues are depicted above the locus and transcription factors with demonstrated binding to the locus are listed below. Numbering is from the start of exon 1b. HS, Hypersensitive site; FP, footprint; RE, response element.
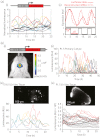
Fig. 3
Transcription dynamics of the human prolactin gene. (
a
) Transcription activity of the human prolactin gene in the somatolactotroph GH3 cell line. Data are from Harper et al. (87). (i) Microscopic measurement of luciferase expression in individual GH3 cells shows that prolactin expression is highly dynamic with pulses of expression. (ii) Luciferase data were used to calculate mRNA expression dynamics of the prolactin gene by taking into account mRNA and protein degradation rates. These data were used to predict ‘on’ and ‘off’ transcription times using a binary switch model. A minimum ‘off’ time of approximately 3 h indicated the presence of a transcriptional refractory period. (
b
) A transgenic rat model with the luciferase gene controlled by the human prolactin locus has enabled investigations of transcription activity in pituitary tissue and primary cells. (i) In vivo imaging of luciferase expression in the pituitary using an IVIS Spectrum (Caliper Life Sciences, Hopkinton, MA, USA). Data from Semprini et al. (15). Technological developments may enable measurements of transcription activity in vivo in real-time in the future. (ii–iv) Luciferase expression reveals marked differences in transcription activity in dissociated cells (ii) and cells maintained within their native tissue environment (iv) and between cells from different stages of development (iii versus iv). Each trace in the graphs represents a single cell. Images of luciferase expression within pituitary tissue are shown above corresponding single cell luciferase expression data. Data from Featherstone et al. (103).
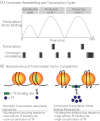
Fig. 4
Chromatin influences on transcription dynamics. (
a
) Chromatin remodelling and transcription factor recruitment define cycles of transcription activity, demonstrated at the pS2 promoter. Active transcription was defined by the presence of phosphorylated RNA polymerase II. Adapted from Metivier et al. (98). (
b
) Nucleosome positioning is plastic and may compete with transcription factors for DNA occupancy affecting transcriptional stochasticity. Factors influencing the equilibrium between nucleosome and transcription factor occupancy include sequence preference for nucleosomes, the affinity of the transcription factor binding site and transcription factor concentration.
Similar articles
- Dynamic analysis of stochastic transcription cycles.
Harper CV, Finkenstädt B, Woodcock DJ, Friedrichsen S, Semprini S, Ashall L, Spiller DG, Mullins JJ, Rand DA, Davis JR, White MR. Harper CV, et al. PLoS Biol. 2011 Apr;9(4):e1000607. doi: 10.1371/journal.pbio.1000607. Epub 2011 Apr 12. PLoS Biol. 2011. PMID: 21532732 Free PMC article. - A family of POU-domain and Pit-1 tissue-specific transcription factors in pituitary and neuroendocrine development.
Ingraham HA, Albert VR, Chen RP, Crenshaw 3d EB, Elsholtz HP, He X, Kapiloff MS, Mangalam HJ, Swanson LW, Treacy MN, et al. Ingraham HA, et al. Annu Rev Physiol. 1990;52:773-91. doi: 10.1146/annurev.ph.52.030190.004013. Annu Rev Physiol. 1990. PMID: 2184776 Review. - Calcium dynamics and chromatin remodelling underlie heterogeneity in prolactin transcription.
Harper CV, McNamara AV, Spiller DG, Charnock JC, White MRH, Davis JRE. Harper CV, et al. J Mol Endocrinol. 2021 Jan;66(1):59-69. doi: 10.1530/JME-20-0223. J Mol Endocrinol. 2021. PMID: 33112804 Free PMC article. - The rat prolactin gene: a target for tissue-specific and hormone-dependent transcription factors.
Gourdji D, Laverrière JN. Gourdji D, et al. Mol Cell Endocrinol. 1994 Apr;100(1-2):133-42. doi: 10.1016/0303-7207(94)90292-5. Mol Cell Endocrinol. 1994. PMID: 7914498 Review. No abstract available.
Cited by
- A stochastic transcriptional switch model for single cell imaging data.
Hey KL, Momiji H, Featherstone K, Davis JR, White MR, Rand DA, Finkenstädt B. Hey KL, et al. Biostatistics. 2015 Oct;16(4):655-69. doi: 10.1093/biostatistics/kxv010. Epub 2015 Mar 26. Biostatistics. 2015. PMID: 25819987 Free PMC article. - Prolactin: A Mammalian Stress Hormone and Its Role in Cutaneous Pathophysiology.
Langan EA. Langan EA. Int J Mol Sci. 2024 Jun 28;25(13):7100. doi: 10.3390/ijms25137100. Int J Mol Sci. 2024. PMID: 39000207 Free PMC article. Review. - Spatially coordinated dynamic gene transcription in living pituitary tissue.
Featherstone K, Hey K, Momiji H, McNamara AV, Patist AL, Woodburn J, Spiller DG, Christian HC, McNeilly AS, Mullins JJ, Finkenstädt BF, Rand DA, White MR, Davis JR. Featherstone K, et al. Elife. 2016 Feb 1;5:e08494. doi: 10.7554/eLife.08494. Elife. 2016. PMID: 26828110 Free PMC article. - Association Between BDNF Gene Variant Rs6265 and the Severity of Depression in Antidepressant Treatment-Free Depressed Patients.
Losenkov IS, Mulder NJV, Levchuk LA, Vyalova NM, Loonen AJM, Bosker FJ, Simutkin GG, Boiko AS, Bokhan NA, Wilffert B, Hak E, Schmidt AF, Ivanova SA. Losenkov IS, et al. Front Psychiatry. 2020 Feb 12;11:38. doi: 10.3389/fpsyt.2020.00038. eCollection 2020. Front Psychiatry. 2020. PMID: 32116853 Free PMC article. - Prolactin decreases LPS-induced inflammatory cytokines by inhibiting TLR-4/NFκB signaling in the human placenta.
Olmos-Ortiz A, Déciga-García M, Preciado-Martínez E, Bermejo-Martínez L, Flores-Espinosa P, Mancilla-Herrera I, Irles C, Helguera-Repetto AC, Quesada-Reyna B, Goffin V, Díaz L, Zaga-Clavellina V. Olmos-Ortiz A, et al. Mol Hum Reprod. 2019 Oct 28;25(10):660-667. doi: 10.1093/molehr/gaz038. Mol Hum Reprod. 2019. PMID: 31263869 Free PMC article.
References
- Walker JJ, Terry JR, Tsaneva-Atanasova K, Armstrong SP, McArdle CA, Lightman SL. Encoding and decoding mechanisms of pulsatile hormone secretion. J Neuroendocrinol. 2010;22:1226–1238. - PubMed
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources