Associations with early-life socio-economic position in adult DNA methylation - PubMed (original) (raw)
Associations with early-life socio-economic position in adult DNA methylation
Nada Borghol et al. Int J Epidemiol. 2012 Feb.
Abstract
Background: Disadvantaged socio-economic position (SEP) in childhood is associated with increased adult mortality and morbidity. We aimed to establish whether childhood SEP was associated with differential methylation of adult DNA.
Methods: Forty adult males from the 1958 British Birth Cohort Study were selected from SEP extremes in both early childhood and mid-adulthood. We performed genome-wide methylation analysis on blood DNA taken at 45 years using MeDIP (methylated DNA immunoprecipitation). We mapped in triplicate the methylation state of promoters of approximately 20,000 genes and 400 microRNAs. Probe methylation scores were averaged across triplicates and differential methylation between groups of individuals was determined. Differentially methylated promoter sites of selected genes were validated using pyrosequencing of bisulfite-converted DNA.
Results: Variably methylated probes (9112 from n = 223,359 on the microarray) corresponded to 6176 gene promoters with at least one variable probe. Unsupervised hierarchical clustering of probes obtained from the 500 most variable promoters revealed a cluster enriched with high SEP individuals confirming that SEP differences contribute to overall epigenetic variation. Methylation levels for 1252 gene promoters were associated with childhood SEP vs 545 promoters for adulthood SEP. Functionally, associations with childhood SEP appear in promoters of genes enriched in key cell signalling pathways. The differentially methylated promoters associated with SEP cluster in megabase-sized regions of the genome.
Conclusions: Adult blood DNA methylation profiles show more associations with childhood SEP than adult SEP. Organization of these associations across the genome suggests a well-defined epigenetic pattern linked to early socio-economic environment.
Figures
Figure 1
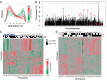
Whole-blood promoter methylation in a variable SEP population. (A) Distributions of promoter methylation levels by published expression level. Genes are divided into 20 levels by whole-blood expression percentiles (0–5, 5–10, … , 95–100) based on publicly available expression data. Shown are the distributions of methylation levels for each expression percentile. The distributions show that genes with low or no expression (represented in green) tend to have highly methylated promoters, whereas genes with high expression (represented in red) tend to have little or no promoter methylation. (B) Distribution of variably methylated promoters. Distribution of variably methylated promoters across the genome shown as percentage of promoters in each region. Downward pointing arrows indicate regions enriched with variably methylated promoters, and upward pointing arrows indicate regions with little variability (FDR < 0.05). The dashed grey line shows the expected percentage of variably methylated promoters. (C) Most variably methylated promoters. Heatmap showing methylation scores of the 500 most variably methylated promoters (rows) across all 40 individuals (columns). Each promoter is represented by its most variable probe. Blackened squares above the columns denote high SEP in childhood or adulthood. The cluster highlighted in blue contains a significant number of high childhood and adulthood SEP individuals (hypergeometric; P ≤ 0.009). Each probe represents a distinct promoter (i.e. for promoters with multiple highly variable probes, only the most variable was included). (D) Differentially methylated promoters associated with childhood SEP. The heatmap shows the methylation scores of the 500 probes most significantly associated with childhood SEP with the following qualifications: each probe satisfied the individual requirements to be called differentially methylated, each belonged to a promoter called differentially methylated, and no pair of probes belonged to the same promoter
Figure 2
Mapping of the state of methylation of three protocadherin genes by pyrosequencing. Effect size (_P_-value) for three protocadherin promoters were as follows: [effect = log(mean probe intensity in high child SEP/mean probe intensity in low child SEP)] PCDHB4 effect = −0.47, _P_-value < 0.00114, _q_-value < 0.015; PCDHB3 effect = −0.34, _P_-value < 0.013, _q_-value < 0.023; PCDHGA11 effect = −0.53, _P_-value < 0.0051, _q_-value < 0.034. In the figure, the upper panel is a UCSC browser genomic display showing mean DNA methylation differences between high and low childhood SEP and the significant differential probe as detected by microarray analysis (A, B and C). Grey bars correspond to probes more methylated in the low childhood SEP group. The chromosomal location of each analysed region is indicated (hg18: human genome 18 assembly). CpG sites are annotated relative to the transcription start site of each gene. Each circle represents one CpG site. The grey box indicates the probe significantly different as shown by the microarrays data. The bar graphs show the average methylation levels of the CpG sites within the high and low childhood SEP groups as determined by pyrosequencing. Error bars show the SEM (*P < 0.05). (A) Methylation profile of the PCDHB4 promoter in childhood high and low SEP groups. In the PCDHB4 promoter, five CpG sites were analyzed by pyrosequencing (high SEP n = 17, low SEP n = 16). On average, all of the CpG sites have higher methylation in the low SEP group (P = 0.0043, Stouffer) with CG7 showing a significant difference (P = 0.018, _t_-test). The lower bar graph shows pyrosequencing average methylation levels for the same five CpGs in additional 30 participants (high SEP n = 13, low SEP n = 17). All five CpG sites are more methylated in the low SEP group with CG7 significantly different (P = 0.011, _t_-test). (B) Methylation profile of the PCDHB3 promoter in childhood high and low SEP groups. In the promoter of the PCDHB3 gene, six CpG sites were analysed by pyrosequencing (high n = 14, low n = 19). Though all CpG sites have higher methylation in the low SEP group, only CG4 is significantly different (P = 0.0049, _t_-test). (C) Methylation profile of the PCDHGA11 promoter in childhood high and low SEP groups. In the promoter of the PCDHGA11 gene, four CpG sites were analysed by pyrosequencing (high n = 16, low n = 20). CG2 shows significantly higher methylation in the low childhood SEP group (P = 0.010, _t_-test)
Figure 3
Megabase co-clustering of differential methylation. (A) Co-clustering of differentially methylated promoters across megabases of DNA. Positive values (black bars) indicate increased methylation in high childhood SEP compared with low childhood SEP and negative values (grey bars) indicate the opposite. With the exception of one region next to the chromosome centromere (highlighted), chromosome 19 promoters are generally more methylated in high childhood SEP than in low childhood SEP. This is in contrast to adulthood SEP where the methylation differences are not significant. (B) Co-clustering of differentially methylated promoters with common function across megabases of DNA. Unlike chromosome 19, chromosome 11 has more directional variation with respect to childhood SEP. We highlight a region of 1 million bases that is consistently more methylated in low childhood SEP and contains about 40 genes, all olfactory receptor genes. Shaded vertical bars identify 500-kb regions of significant difference between childhood SEP groups (FDR ≤ 0.025). (C) Co-clustering of differential methylation with the protocadherins. Shaded is a nearly 1-Mb region containing mainly protocadherins whose promoters are consistently more methylated in low childhood SEP than in high. Most of this region was found to be consistently hypermethylated in breast tumours. We observe a similar but much weaker and inverted methylation difference between low and high adulthood SEP
Figure 4
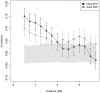
Methylation dependencies across megabases. Shown are correlations of methylation differences from 500-kb regions at various distances apart. The solid grey region contains the 95% CI, and error bars contain the 95% CI for correlation values
Comment in
- Commentary: The seven plagues of epigenetic epidemiology.
Heijmans BT, Mill J. Heijmans BT, et al. Int J Epidemiol. 2012 Feb;41(1):74-8. doi: 10.1093/ije/dyr225. Epub 2012 Jan 23. Int J Epidemiol. 2012. PMID: 22269254 Free PMC article. No abstract available.
Similar articles
- Childhood abuse is associated with methylation of multiple loci in adult DNA.
Suderman M, Borghol N, Pappas JJ, Pinto Pereira SM, Pembrey M, Hertzman C, Power C, Szyf M. Suderman M, et al. BMC Med Genomics. 2014 Mar 11;7:13. doi: 10.1186/1755-8794-7-13. BMC Med Genomics. 2014. PMID: 24618023 Free PMC article. - Socioeconomic position during pregnancy and DNA methylation signatures at three stages across early life: epigenome-wide association studies in the ALSPAC birth cohort.
Alfano R, Guida F, Galobardes B, Chadeau-Hyam M, Delpierre C, Ghantous A, Henderson J, Herceg Z, Jain P, Nawrot TS, Relton C, Vineis P, Castagné R, Plusquin M. Alfano R, et al. Int J Epidemiol. 2019 Feb 1;48(1):30-44. doi: 10.1093/ije/dyy259. Int J Epidemiol. 2019. PMID: 30590607 Free PMC article. - Lymphoblastoid cell lines reveal associations of adult DNA methylation with childhood and current adversity that are distinct from whole blood associations.
Suderman M, Pappas JJ, Borghol N, Buxton JL, McArdle WL, Ring SM, Hertzman C, Power C, Szyf M, Pembrey M. Suderman M, et al. Int J Epidemiol. 2015 Aug;44(4):1331-40. doi: 10.1093/ije/dyv168. Epub 2015 Sep 8. Int J Epidemiol. 2015. PMID: 26351305 - Associations between childhood socioeconomic position and adulthood obesity.
Senese LC, Almeida ND, Fath AK, Smith BT, Loucks EB. Senese LC, et al. Epidemiol Rev. 2009;31:21-51. doi: 10.1093/epirev/mxp006. Epub 2009 Jul 31. Epidemiol Rev. 2009. PMID: 19648176 Free PMC article. Review. - DNA methylation signatures as biomarkers of socioeconomic position.
Rajaprakash M, Dean LT, Palmore M, Johnson SB, Kaufman J, Fallin DM, Ladd-Acosta C. Rajaprakash M, et al. Environ Epigenet. 2022 Dec 14;9(1):dvac027. doi: 10.1093/eep/dvac027. eCollection 2023. Environ Epigenet. 2022. PMID: 36694711 Free PMC article. Review.
Cited by
- Transcriptional modulation of the developing immune system by early life social adversity.
Cole SW, Conti G, Arevalo JM, Ruggiero AM, Heckman JJ, Suomi SJ. Cole SW, et al. Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):20578-83. doi: 10.1073/pnas.1218253109. Epub 2012 Nov 26. Proc Natl Acad Sci U S A. 2012. PMID: 23184974 Free PMC article. - Mouse Social Network Dynamics and Community Structure are Associated with Plasticity-Related Brain Gene Expression.
Williamson CM, Franks B, Curley JP. Williamson CM, et al. Front Behav Neurosci. 2016 Aug 4;10:152. doi: 10.3389/fnbeh.2016.00152. eCollection 2016. Front Behav Neurosci. 2016. PMID: 27540359 Free PMC article. - Metabolic, hormonal and immunological associations with global DNA methylation among postmenopausal women.
Ulrich CM, Toriola AT, Koepl LM, Sandifer T, Poole EM, Duggan C, McTiernan A, Issa JP. Ulrich CM, et al. Epigenetics. 2012 Sep;7(9):1020-8. doi: 10.4161/epi.21464. Epub 2012 Aug 7. Epigenetics. 2012. PMID: 22869041 Free PMC article. - MAOA promoter methylation and susceptibility to carotid atherosclerosis: role of familial factors in a monozygotic twin sample.
Zhao J, Forsberg CW, Goldberg J, Smith NL, Vaccarino V. Zhao J, et al. BMC Med Genet. 2012 Nov 2;13:100. doi: 10.1186/1471-2350-13-100. BMC Med Genet. 2012. PMID: 23116433 Free PMC article. - Commentary: The seven plagues of epigenetic epidemiology.
Heijmans BT, Mill J. Heijmans BT, et al. Int J Epidemiol. 2012 Feb;41(1):74-8. doi: 10.1093/ije/dyr225. Epub 2012 Jan 23. Int J Epidemiol. 2012. PMID: 22269254 Free PMC article. No abstract available.
References
- Acheson D. Inequalities in Health: Report of an Independent Inquiry. London: HMSO; 1998.
- Berkman LF, Kawachi IO. Social Epidemiology. New York: Oxford University Press; 2000.
- WHO Commission on Social Determinants of Health, World Health Organization. Closing the Gap in a Generation: Health Equity through Action on the Social Determinants of Health: Commission on Social Determinants of Health Final Report: Executive Summary. Geneva, Switzerland: World Health Organization, Commission on Social Determinants of Health; 2008.
- Galobardes B, Lynch JW, Davey Smith G. Childhood socioeconomic circumstances and cause-specific mortality in adulthood: systematic review and interpretation. Epidemiol Rev. 2004;26:7–21. - PubMed
- Gluckman PD, Hanson MA, Bateson P, et al. Towards a new developmental synthesis: adaptive developmental plasticity and human disease. Lancet. 2009;373:1654–57. - PubMed
Publication types
MeSH terms
Grants and funding
- G1001799/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- CAPMC/ CIHR/Canada
- DH_/Department of Health/United Kingdom
- G0400546/MRC_/Medical Research Council/United Kingdom
- G0000934/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases