Sequence evolution correlates with structural dynamics - PubMed (original) (raw)
Sequence evolution correlates with structural dynamics
Ying Liu et al. Mol Biol Evol. 2012 Sep.
Abstract
Biochemical activity and core stability are essential properties of proteins, maintained usually by conserved amino acids. Structural dynamics emerged in recent years as another essential aspect of protein functionality. Structural dynamics enable the adaptation of the protein to binding substrates and to undergo allosteric transitions, while maintaining the native fold. Key residues that mediate structural dynamics would thus be expected to be conserved or exhibit coevolutionary patterns at least. Yet, the correlation between sequence evolution and structural dynamics is yet to be established. With recent advances in efficient characterization of structural dynamics, we are now in a position to perform a systematic analysis. In the present study, a set of 34 enzymes representing various folds and functional classes is analyzed using information theory and elastic network models. Our analysis shows that the structural regions distinguished by their coevolution propensity as well as high mobility are predisposed to serve as substrate recognition sites, whereas residues acting as global hinges during collective dynamics are often supported by conserved residues. We propose a mobility scale for different types of amino acids, which tends to vary inversely with amino acid conservation. Our findings suggest the balance between physical adaptability (enabled by structure-encoded motions) and chemical specificity (conferred by correlated amino acid substitutions) underlies the selection of a relatively small set of versatile folds by proteins.
Figures
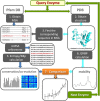
Fig. 1.
Workflow of the study. For each query enzyme in the data set, we retrieve the structure from the PDB and the MSA from Pfam database. These are used as input for 1) GNM evaluation of residue mobilities (right branch) and 2) generation of conservation profile and coevolution maps (left branch), respectively. Comparison of the outputs shows that sequence entropy is accompanied by conformational mobility (enhanced dynamics), correlated mutations exhibit a broad range of mobilities depending on the type of underlying evolutionary pressure, and conserved sites are practically immobile. Statistically significant results are obtained by compiling the outputs for 34 enzymes.
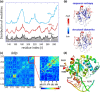
Fig. 2.
An illustrative example: comparative analysis of residue conservation, conformational mobility, and coevolutionary patterns for UDG. (a) Mobility and conservation profiles as a function of residue index. Blue, red, and black curves represent the mobility profiles <_M_ _i_>|_m_1, <_M_ _i_>|_m_2, and <_M_ _i_>|N − 1 (or MSFs) computed using the GNM. The curves are shifted vertically for clarity. The bars represent the information entropy derived from 1599 Pfam sequences (
supplementary table S1
,
Supplementary Material
online). Results are shown for the structurally resolved residues 131 ≤ i ≤ 292 that are fully represented in the MSA. (b) Comparison of conservation (upper) and mobility (lower) profiles using color-coded ribbon diagrams. (c) MIp map for the UDG family (see
supplementary fig. S2
,
Supplementary Material
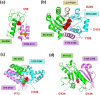
online, for the corresponding MI map). The magnified portion refers to the DNA-binding region of UDG. Highest signals are detected at M131-I134, P163, V164, I181-F184, C281, and H283. (d) Location of residues distinguished by high MIp values at DNA-binding site. The diagram is color coded based on the crystallographic B factors (red/blue: most/least mobile) reported for UDG.
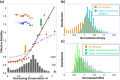
Fig. 3.
Relationship between structural dynamics and sequence evolution properties. (a) Effective mobility as a function of sequence conservation, based on softest modes (red circles) or N − 1 modes (open circles) computed for all residues in the data set of 34 enzymes. The curves are the weighted least square fits to computed data, with respective correlation coefficients of 0.90 and 0.95. The number distribution of residues in different entropy intervals is shown by the gray bars (right ordinate). Entries with Si > 2 are merged in the last bin. Arrows delimit distinctive mobility versus conservation regimes. (b) Sequence entropy distribution for all residues (orange) and a subset distinguished by their high coevolution propensities (cyan). (c) Mobility histograms for three groups of residues, as labeled. Respective mean values and variances are 1.00 ± 0.134, 0.79 ± 0.059, and 1.06 ± 0.127.
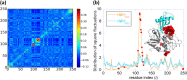
Fig. 4.
Sequence coevolution and high mobility properties at the ligand recognition site of procathepsin B catalytic domain. (a) MI map, highlighting (in red) the coevolving amino acid pairs. Residues corresponding to the top 0.05% MIp values (N47, A48, S65, M66, I105, C108, N113-P118, T120-G123, T125, A248, and G249) are indicated by squares on the <Mieff>|m1 curve, color. They are shown by spheres in the ribbon diagram for the complex formed with stefin A (cyan). (b) Global mobility profile (orange) and MSF distribution of residues (cyan) for procathepsin B. The residues distinguished in panel a by their coevolutionary propensities are shown by red spheres in the ribbon diagram of the protein (gray/red). Note the close neighborhood of this region to the binding site of the substrate stefin A (cyan).
Fig. 5.
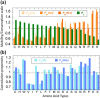
Mobility, conservation, and coevolution propensities of amino acids. (a) Distributions of amino acids within the subsets composed of highly conserved (_C_-) (green bars) and highly mobile (_M_-) sites (light-to-dark orange bars, based on m1, m2, or N − 1 modes, as labeled). The bars represent the propensities with respect to those expected a priori based on the frequency of occurrence of the particular amino acid types in the data set. (b) Coevolution propensities of amino acids based on MI (light blue) and MIp (dark blue) values, as labeled. Amino acid types (shown by one-letter codes) are listed in the order of decreasing entropy in both panels.
Fig. 6.
Conserved sites distinguished by minimal fluctuations in global modes, despite moderate-to-high exposure to solvent. The figure illustrates four cases: (a) Staphyloccocal nuclease (PDB id: 1kab), (b) exonuclease III (PDB id: 1ako), (c) phospholipase C (PDB id: 2ffz), and (d) dehydrofolate reductase (PDB id: 3cd2). The labeled residues displayed in red space-filling representations simultaneously belong to the _C_- and _H_-subsets (of highly conserved and dynamically restrained residues) but not to the _B_-subset (of most buried residues). The identities of these residues and substructures whose collective dynamics they delimit are indicated by the labels (color coded after the substructures). The orange, space-filling residues in panel a illustrate a pair of residues that are highly conserved and buried (but globally moving as part of the violet substructure).
Similar articles
- Protein promiscuity: drug resistance and native functions--HIV-1 case.
Fernández A, Tawfik DS, Berkhout B, Sanders R, Kloczkowski A, Sen T, Jernigan B. Fernández A, et al. J Biomol Struct Dyn. 2005 Jun;22(6):615-24. doi: 10.1080/07391102.2005.10531228. J Biomol Struct Dyn. 2005. PMID: 15842167 - Evolution of function in protein superfamilies, from a structural perspective.
Todd AE, Orengo CA, Thornton JM. Todd AE, et al. J Mol Biol. 2001 Apr 6;307(4):1113-43. doi: 10.1006/jmbi.2001.4513. J Mol Biol. 2001. PMID: 11286560 - Improvement in protein functional site prediction by distinguishing structural and functional constraints on protein family evolution using computational design.
Cheng G, Qian B, Samudrala R, Baker D. Cheng G, et al. Nucleic Acids Res. 2005 Oct 13;33(18):5861-7. doi: 10.1093/nar/gki894. Print 2005. Nucleic Acids Res. 2005. PMID: 16224101 Free PMC article. - A global analysis of function and conservation of catalytic residues in enzymes.
Ribeiro AJM, Tyzack JD, Borkakoti N, Holliday GL, Thornton JM. Ribeiro AJM, et al. J Biol Chem. 2020 Jan 10;295(2):314-324. doi: 10.1074/jbc.REV119.006289. Epub 2019 Dec 3. J Biol Chem. 2020. PMID: 31796628 Free PMC article. Review. - Structural and functional restraints in the evolution of protein families and superfamilies.
Gong S, Worth CL, Bickerton GR, Lee S, Tanramluk D, Blundell TL. Gong S, et al. Biochem Soc Trans. 2009 Aug;37(Pt 4):727-33. doi: 10.1042/BST0370727. Biochem Soc Trans. 2009. PMID: 19614584 Review.
Cited by
- Structural bioinformatics enhances the interpretation of somatic mutations in KDM6A found in human cancers.
Chi YI, Stodola TJ, De Assuncao TM, Leverence EN, Smith BC, Volkman BF, Mathison AJ, Lomberk G, Zimmermann MT, Urrutia R. Chi YI, et al. Comput Struct Biotechnol J. 2022 Apr 28;20:2200-2211. doi: 10.1016/j.csbj.2022.04.028. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35615018 Free PMC article. - Identification of potential CD8+ epitopes in pp62 polyprotein of African swine fever virus using computational immunology.
Galicia MLC, Morales DJM, Pogado PGB, Quebrado AL, Herrera-Ong LR. Galicia MLC, et al. BioTechnologia (Pozn). 2023 Sep 25;104(3):221-231. doi: 10.5114/bta.2023.130726. eCollection 2023. BioTechnologia (Pozn). 2023. PMID: 37850118 Free PMC article. - Optimization of Conformational Dynamics in an Epistatic Evolutionary Trajectory.
González MM, Abriata LA, Tomatis PE, Vila AJ. González MM, et al. Mol Biol Evol. 2016 Jul;33(7):1768-76. doi: 10.1093/molbev/msw052. Epub 2016 Mar 15. Mol Biol Evol. 2016. PMID: 26983555 Free PMC article. - Combining protein sequence, structure, and dynamics: A novel approach for functional evolution analysis of PAS domain superfamily.
Dong Z, Zhou H, Tao P. Dong Z, et al. Protein Sci. 2018 Feb;27(2):421-430. doi: 10.1002/pro.3329. Epub 2017 Nov 2. Protein Sci. 2018. PMID: 29052279 Free PMC article. - Allosteric signalling in the outer membrane translocation domain of PapC usher.
Farabella I, Pham T, Henderson NS, Geibel S, Phan G, Thanassi DG, Delcour AH, Waksman G, Topf M. Farabella I, et al. Elife. 2014 Oct 28;3:e03532. doi: 10.7554/eLife.03532. Elife. 2014. PMID: 25271373 Free PMC article.
References
- Atchley WR, Wollenberg KR, Fitch WM, Terhalle W, Dress AW. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol Biol Evol. 2000;17:164–178. - PubMed
- Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–181. - PubMed
- Betts MJ, Russell RB. Amino-acid properties and consequences of substitutions. In: Barnes MR, editor. Bioinformatics for geneticists: a bioinformatics primer for the analysis of genetic data. Chichester (UK): John Wiley & Sons Ltd; 2007. pp. 311–342.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources