Microbes inside--from diversity to function: the case of Akkermansia - PubMed (original) (raw)
Review
Microbes inside--from diversity to function: the case of Akkermansia
Clara Belzer et al. ISME J. 2012 Aug.
Abstract
The human intestinal tract is colonized by a myriad of microbes that have developed intimate interactions with the host. In healthy individuals, this complex ecosystem remains stable and resilient to stressors. There is significant attention on the understanding of the composition and function of this intestinal microbiota in health and disease. Current developments in metaomics and systems biology approaches allow to probe the functional potential and activity of the intestinal microbiota. However, all these approaches inherently suffer from the fact that the information on macromolecules (DNA, RNA and protein) is collected at the ecosystem level. Similarly, all physiological and other information collected from isolated strains relates to pure cultures grown in vitro or in gnotobiotic systems. It is essential to integrate these two worlds of predominantly chemistry and biology by linking the molecules to the cells. Here, we will address the integration of omics- and culture-based approaches with the complexity of the human intestinal microbiota in mind and the mucus-degrading bacteria Akkermansia spp. as a paradigm.
Figures
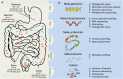
Figure 1
From diversity to function. (a) Schematic outline of the human digestive tract with its characteristics and microbiota. The stomach is characterized by low pH conditions and a short retention time and colonization by bacteria is minimal. The small intestine (duodenum, jejunum and proximal ileum) has a short retention time (2–6 h) and high pH, the residing microflora is exposed to secreted bile salts and pancreatic juices at the proximal part. The lower digestive tract, comprising the terminal ileum and colon, is in contrast characterized by a longer retention time, neutral pH and is the most densely populated by microorganisms. (b) Overview of the major ‘omics' approaches using DNA, RNA, protein or culture-based techniques.
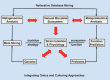
Figure 2
Integrating omics and culturing approaches. All metagenomics and other omics approaches suffer from the fact that information on the macromolecules (DNA, RNA or protein) is collected at the ecosystem level. What needs to be done is to integrate these two worlds of predominantly chemistry and biology by linking the molecules to the cells. Red arrows indicate the experimental approaches related to this integration and the other arrows describe other activities and approaches. See text for further explanations.
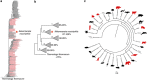
Figure 3
Akkermansia muciniphila is universally distributed in intestinal tracts all over the animal kingdom. (a) Phylogenetic tree indicating the position of A. muciniphila among selected full-length 16S rRNA clones from mammalian gut samples. Red colored samples derive from human sources. Thermotoga thermarum is used as an outgroup. The tree was generated using the neighbor joining method. Full details and high-resolution information are provided in Supplementary Figure S1. (b) Schematic representation of the tree in (a) with the five different clades their position and similarity to A. muciniphila. (c) Taxonomic tree of mammals generated using iTol webtool from tree of life project using all available sequences from NCBI (Letunic and Bork). Animal silhouettes indicate single species as a representative of that order. When an animal species from the mammalian orders was positive for _Akkermansia_-like sequences the animal logo belonging to that order is colored red, when it was negative the animal logo is colored gray. No Akkermansia sequences have been reported yet in any of the animals belonging to the mammalian orders depicted in black.
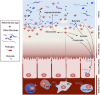
Figure 4
Akkermansia muciniphila activity and interactions in the intestine. Schematic overview of the metabolic activities of A. muciniphila in the gut and the microbiota and host response as a result of A. muciniphila colonization. As a result of mucus degradation, A. muciniphila produces oligosaccharides and SCFAs. These products can stimulate microbiota interactions and host response. Oligosaccharides and acetate stimulate growth and metabolic activity of bacteria that colonize close to the mucus layer. This may provide colonization resistance to pathogenic bacteria that have to cross the mucus layer to reach the intestinal cells. The propionate produced by _Akkermansia_-like bacteria can signal to the host via the Gpr43 receptor and other SCFA may also do the same via Gpr41 (Le Poul et al., 2003; Maslowski et al., 2009). This may trigger a cascade of responses in the host expression machinery and together with other signaling pathways has shown to result in immune stimulation and metabolic signaling in monoassociated germ-free mice (Derrien et al., 2011).
Similar articles
- Genome-Scale Model and Omics Analysis of Metabolic Capacities of Akkermansia muciniphila Reveal a Preferential Mucin-Degrading Lifestyle.
Ottman N, Davids M, Suarez-Diez M, Boeren S, Schaap PJ, Martins Dos Santos VAP, Smidt H, Belzer C, de Vos WM. Ottman N, et al. Appl Environ Microbiol. 2017 Aug 31;83(18):e01014-17. doi: 10.1128/AEM.01014-17. Print 2017 Sep 15. Appl Environ Microbiol. 2017. PMID: 28687644 Free PMC article. - The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes.
van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. van Passel MW, et al. PLoS One. 2011 Mar 3;6(3):e16876. doi: 10.1371/journal.pone.0016876. PLoS One. 2011. PMID: 21390229 Free PMC article. - Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions.
Kolmeder CA, de Been M, Nikkilä J, Ritamo I, Mättö J, Valmu L, Salojärvi J, Palva A, Salonen A, de Vos WM. Kolmeder CA, et al. PLoS One. 2012;7(1):e29913. doi: 10.1371/journal.pone.0029913. Epub 2012 Jan 18. PLoS One. 2012. PMID: 22279554 Free PMC article. - The intestinal environment in health and disease - recent insights on the potential of intestinal bacteria to influence human health.
Possemiers S, Grootaert C, Vermeiren J, Gross G, Marzorati M, Verstraete W, Van de Wiele T. Possemiers S, et al. Curr Pharm Des. 2009;15(18):2051-65. doi: 10.2174/138161209788489159. Curr Pharm Des. 2009. PMID: 19519443 Review. - Impact of the intestinal microbiota on the development of mucosal defense.
Gaskins HR, Croix JA, Nakamura N, Nava GM. Gaskins HR, et al. Clin Infect Dis. 2008 Feb 1;46 Suppl 2:S80-6; discussion S144-51. doi: 10.1086/523336. Clin Infect Dis. 2008. PMID: 18181729 Review.
Cited by
- Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity.
Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Everard A, et al. Proc Natl Acad Sci U S A. 2013 May 28;110(22):9066-71. doi: 10.1073/pnas.1219451110. Epub 2013 May 13. Proc Natl Acad Sci U S A. 2013. PMID: 23671105 Free PMC article. - Beneficial effects of mung bean seed coat on the prevention of high-fat diet-induced obesity and the modulation of gut microbiota in mice.
Hou D, Zhao Q, Yousaf L, Xue Y, Shen Q. Hou D, et al. Eur J Nutr. 2021 Jun;60(4):2029-2045. doi: 10.1007/s00394-020-02395-x. Epub 2020 Oct 1. Eur J Nutr. 2021. PMID: 33005980 - Standard diet and animal source influence hippocampal spatial reference learning and memory in congenic C57BL/6J mice.
Hart DW, Sherman MA, Kim M, Pelzel R, Brown JL, Lesné SE. Hart DW, et al. Res Sq [Preprint]. 2024 Jul 16:rs.3.rs-4582616. doi: 10.21203/rs.3.rs-4582616/v1. Res Sq. 2024. PMID: 39070656 Free PMC article. Preprint. - Dynamics of Gut Microbiota and Clinical Variables after Ketogenic and Mediterranean Diets in Drug-Naïve Patients with Type 2 Diabetes Mellitus and Obesity.
Deledda A, Palmas V, Heidrich V, Fosci M, Lombardo M, Cambarau G, Lai A, Melis M, Loi E, Loviselli A, Manzin A, Velluzzi F. Deledda A, et al. Metabolites. 2022 Nov 10;12(11):1092. doi: 10.3390/metabo12111092. Metabolites. 2022. PMID: 36355175 Free PMC article. - Rumen microbiota of indigenous and introduced ruminants and their adaptation to the Qinghai-Tibetan plateau.
Li B, Jia G, Wen D, Zhao X, Zhang J, Xu Q, Zhao X, Jiang N, Liu Z, Wang Y. Li B, et al. Front Microbiol. 2022 Oct 10;13:1027138. doi: 10.3389/fmicb.2022.1027138. eCollection 2022. Front Microbiol. 2022. PMID: 36299720 Free PMC article. Review.
References
- Akkermans ADL, van Elsas JD, de Bruijn FJ. Molecular Microbial Ecology Manual. Kluwer Academic Publishers: Dordrecht, The Netherlands; 1996.
- Booijink CC, El-Aidy S, Rajilic-Stojanovic M, Heilig HG, Troost FJ, Smidt H, et al. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12:3213–3227. - PubMed
- Boschker HTS, Nold SC, Wellsbury P, Bos D, de Graaf W, Pel R, et al. CappenbergDirect linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature. 1998;392:801–805.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous