A novel STAT1 mutation associated with disseminated mycobacterial disease - PubMed (original) (raw)
Case Reports
. 2012 Aug;32(4):681-689.
doi: 10.1007/s10875-012-9659-2. Epub 2012 Feb 29.
Hannelore I Bax # 1 3, Amy P Hsu 1, Ervand Kristosturyan 1, Joseph Pechacek 1, Prabha Chandrasekaran 1, Michelle L Paulson 4, Dalton L Dias 1, Christine Spalding 1, Gulbu Uzel 1, Li Ding 1, Elizabeth McFarland 5, Steven M Holland 1
Affiliations
- PMID: 22437822
- PMCID: PMC4112946
- DOI: 10.1007/s10875-012-9659-2
Case Reports
A novel STAT1 mutation associated with disseminated mycobacterial disease
Elizabeth P Sampaio et al. J Clin Immunol. 2012 Aug.
Abstract
STAT1 is a key component of Interferon (IFN)-γ and IFN-α signaling and mediates protection against mycobacteria, fungal, viral infections, and cancer. Dominant negative inhibitory as well as gain of function heterozygous STAT1 mutations demonstrate that IFN-γ driven cellular responses need to be tightly regulated to control infections. We describe an autosomal dominant mutation in the SH2 domain of STAT1 that disrupts protein phosphorylation, c.1961T>A (M654K). The mutant allele does not permit STAT1 phosphorylation, and impairs STAT1 phosphorylation of the wild type allele. Protein dimerization is preserved but DNA binding activity, IFN-γ driven GAS-luciferase activity, and expression of IFN-γ target genes are reduced. IFN-α driven ISRE response, but not IFN-α driven GAS response, are preserved when cells are co-transfected with wild type and the mutant STAT1 constructs. M654K exerts a dominant negative effect on IFN-γ related immunity and is recessive for IFN-α induced immune function.
Figures
Fig. 1
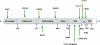
STAT1 dominant negative and loss of function mutations. The N-terminal domain, coiled-coil domain, DNA binding domain, linker domain, SH2 domain, tail segment domain (TS), and transactivation domain (TAD) are represented; Y701, site of tyrosine phosphorylation. The dominant negative mutants are indicated above the protein and all recessive or loss of function mutations are below the protein. M654K is shown in red.
Fig. 2
Dimerization and activation of STAT1 protein. a For evaluation of STAT1 dimerization, U3A cells were transfected with WT and patient M654K STAT1 constructs tagged with either Myc or Flag. Proteins obtained from non stimulated (NS) or IFN-γ stimulated (400 IU/ml) cells were co-precipitated with antibodies against Myc or Flag and blotted for the detection of Myc or Flag. Blots were stripped and reprobed with anti-total STAT1 antibody; b For Western blot analysis, whole cell lysates obtained from transfected U3A cells stimulated or not (NS) with IFN-γ were blotted with anti-phosphorylated STAT1 (pSTAT1). Blots were stripped and reprobed with anti-STAT1 antibody; c Evaluation of U3A cells by flow cytometry confirmed the absent phosphorylation in the M654K transfected cells in response to IFN-γ. Lower phosphorylation is sustained in the co-transfected cells (M654K+WT). Histograms from one representative experiment are presented (right panel). The bars represent mean fold change (±SEM) in mean fluorescence value (MFV) observed in the stimulated cells relative to the unstimulated controls (uns) (n=3). **p<0.01; ***p<0.001 when compared to WT; d EBV transformed B cells obtained from healthy donors and from the STAT1 deficient patient (M654K) were stimulated with IFN-γ or IFN-α. Lysates were blotted with anti-phospho STAT1. Blots were treated as described above and are representative of three individual experiments. NS = non stimulated; e Flow cytometry assayed in EBV-B cells from patients (M654K STAT1 deficient and one patient with complete IFNGR1 deficiency) and two healthy donors. Histograms are included for each experimental condition (non stimulated, NS; IFN-γ 400 IU/ml; IFN-α 1,000 IU/ml). One representative experiment out of three is presented.
Fig. 2
Dimerization and activation of STAT1 protein. a For evaluation of STAT1 dimerization, U3A cells were transfected with WT and patient M654K STAT1 constructs tagged with either Myc or Flag. Proteins obtained from non stimulated (NS) or IFN-γ stimulated (400 IU/ml) cells were co-precipitated with antibodies against Myc or Flag and blotted for the detection of Myc or Flag. Blots were stripped and reprobed with anti-total STAT1 antibody; b For Western blot analysis, whole cell lysates obtained from transfected U3A cells stimulated or not (NS) with IFN-γ were blotted with anti-phosphorylated STAT1 (pSTAT1). Blots were stripped and reprobed with anti-STAT1 antibody; c Evaluation of U3A cells by flow cytometry confirmed the absent phosphorylation in the M654K transfected cells in response to IFN-γ. Lower phosphorylation is sustained in the co-transfected cells (M654K+WT). Histograms from one representative experiment are presented (right panel). The bars represent mean fold change (±SEM) in mean fluorescence value (MFV) observed in the stimulated cells relative to the unstimulated controls (uns) (n=3). **p<0.01; ***p<0.001 when compared to WT; d EBV transformed B cells obtained from healthy donors and from the STAT1 deficient patient (M654K) were stimulated with IFN-γ or IFN-α. Lysates were blotted with anti-phospho STAT1. Blots were treated as described above and are representative of three individual experiments. NS = non stimulated; e Flow cytometry assayed in EBV-B cells from patients (M654K STAT1 deficient and one patient with complete IFNGR1 deficiency) and two healthy donors. Histograms are included for each experimental condition (non stimulated, NS; IFN-γ 400 IU/ml; IFN-α 1,000 IU/ml). One representative experiment out of three is presented.
Fig. 2
Dimerization and activation of STAT1 protein. a For evaluation of STAT1 dimerization, U3A cells were transfected with WT and patient M654K STAT1 constructs tagged with either Myc or Flag. Proteins obtained from non stimulated (NS) or IFN-γ stimulated (400 IU/ml) cells were co-precipitated with antibodies against Myc or Flag and blotted for the detection of Myc or Flag. Blots were stripped and reprobed with anti-total STAT1 antibody; b For Western blot analysis, whole cell lysates obtained from transfected U3A cells stimulated or not (NS) with IFN-γ were blotted with anti-phosphorylated STAT1 (pSTAT1). Blots were stripped and reprobed with anti-STAT1 antibody; c Evaluation of U3A cells by flow cytometry confirmed the absent phosphorylation in the M654K transfected cells in response to IFN-γ. Lower phosphorylation is sustained in the co-transfected cells (M654K+WT). Histograms from one representative experiment are presented (right panel). The bars represent mean fold change (±SEM) in mean fluorescence value (MFV) observed in the stimulated cells relative to the unstimulated controls (uns) (n=3). **p<0.01; ***p<0.001 when compared to WT; d EBV transformed B cells obtained from healthy donors and from the STAT1 deficient patient (M654K) were stimulated with IFN-γ or IFN-α. Lysates were blotted with anti-phospho STAT1. Blots were treated as described above and are representative of three individual experiments. NS = non stimulated; e Flow cytometry assayed in EBV-B cells from patients (M654K STAT1 deficient and one patient with complete IFNGR1 deficiency) and two healthy donors. Histograms are included for each experimental condition (non stimulated, NS; IFN-γ 400 IU/ml; IFN-α 1,000 IU/ml). One representative experiment out of three is presented.
Fig. 3
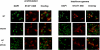
Evaluation of nuclear translocation. Nuclear translocation of the mutant STAT1 proteins was assessed in U3A cells transfected with WT STAT1 or M654K mutant, stimulated or not with IFN-γ (400 IU/ml, 15 min). The distribution of STAT1 in the nucleus was identified by primary staining against total STAT1 followed by a secondary staining with Alexa Fluor 568, together with the nuclear DAPI. Following stimulation, WT and WT+M654K STAT1 but not the M654K alone were detected at the nucleus as shown in the overlay image.
Fig. 4
Effect of STAT1 mutation on DNA binding and transcriptional activity. a STAT1 DNA-binding activity was assayed in nuclear extracts isolated from U3A cells transfected with STAT1 WT, STAT1 mutant (M654K), or STAT1 mutant co-transfected with WT (M654K+WT) and stimulated or not with IFN-γ or IFN-α 1,000 IU/ml (30 min). Data are mean fold increase (± SD) detected in the stimulated cultures relative to the WT non stimulated (NS) from a total of five experiments; b STAT1 expression vectors (STAT1 WT, mutant M654K, M654K+WT) were transiently co-transfected with luciferase GAS reporter plasmids or c with luciferase ISRE reporter into U3A cells. Following stimulation with IFN-γ or IFN-α, cells were harvested and assayed using the dual luciferase reporter assay. Experiments were done in triplicate and results are mean fold increase (± SD) in the stimulated cells relative to the WT non stimulated (n=4).**p<0.01; ***p<0.001 when compared to WT.
Fig. 4
Effect of STAT1 mutation on DNA binding and transcriptional activity. a STAT1 DNA-binding activity was assayed in nuclear extracts isolated from U3A cells transfected with STAT1 WT, STAT1 mutant (M654K), or STAT1 mutant co-transfected with WT (M654K+WT) and stimulated or not with IFN-γ or IFN-α 1,000 IU/ml (30 min). Data are mean fold increase (± SD) detected in the stimulated cultures relative to the WT non stimulated (NS) from a total of five experiments; b STAT1 expression vectors (STAT1 WT, mutant M654K, M654K+WT) were transiently co-transfected with luciferase GAS reporter plasmids or c with luciferase ISRE reporter into U3A cells. Following stimulation with IFN-γ or IFN-α, cells were harvested and assayed using the dual luciferase reporter assay. Experiments were done in triplicate and results are mean fold increase (± SD) in the stimulated cells relative to the WT non stimulated (n=4).**p<0.01; ***p<0.001 when compared to WT.
Fig. 5
For evaluation of STAT1 / STAT2 association, U3A cells were transfected with WT STAT1 or M654K mutant constructs, left non stimulated (NS) or stimulated with IFN-α (1,000 IU/ml) for 30 min when cell lysates were prepared and co-precipitated with anti-STAT1 antibody and blotted for STAT2. Blots were stripped and reprobed with anti-total STAT1. One representative experiment out of two is presented.
Fig. 6
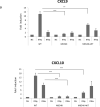
Analysis of gene expression. U3A cells co-transfected with WT and/or patient construct (M654K) were stimulated or not with IFN-γ or IFN-α for 3 h. Expression of IFN target genes, a CXCL9, CXCL10, b MX1, OAS2 was evaluated in the transfected cells by real time PCR. Results are expressed as mean fold induction (± SD) of four individual experiments. *p <0.05; **p < 0.01; ***p < 0.001 when compared to WT.
Fig. 6
Analysis of gene expression. U3A cells co-transfected with WT and/or patient construct (M654K) were stimulated or not with IFN-γ or IFN-α for 3 h. Expression of IFN target genes, a CXCL9, CXCL10, b MX1, OAS2 was evaluated in the transfected cells by real time PCR. Results are expressed as mean fold induction (± SD) of four individual experiments. *p <0.05; **p < 0.01; ***p < 0.001 when compared to WT.
Similar articles
- Dominant-negative STAT1 SH2 domain mutations in unrelated patients with Mendelian susceptibility to mycobacterial disease.
Tsumura M, Okada S, Sakai H, Yasunaga S, Ohtsubo M, Murata T, Obata H, Yasumi T, Kong XF, Abhyankar A, Heike T, Nakahata T, Nishikomori R, Al-Muhsen S, Boisson-Dupuis S, Casanova JL, Alzahrani M, Shehri MA, Elghazali G, Takihara Y, Kobayashi M. Tsumura M, et al. Hum Mutat. 2012 Sep;33(9):1377-87. doi: 10.1002/humu.22113. Epub 2012 Jun 7. Hum Mutat. 2012. PMID: 22573496 Free PMC article. - Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease.
Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, Casrouge A, Yang K, Soudais C, Fieschi C, Santos OF, Bustamante J, Picard C, de Beaucoudrey L, Emile JF, Arkwright PD, Schreiber RD, Rolinck-Werninghaus C, Rösen-Wolff A, Magdorf K, Roesler J, Casanova JL. Chapgier A, et al. PLoS Genet. 2006 Aug 18;2(8):e131. doi: 10.1371/journal.pgen.0020131. PLoS Genet. 2006. PMID: 16934001 Free PMC article. - Heterozygosity for the Y701C STAT1 mutation in a multiplex kindred with multifocal osteomyelitis.
Hirata O, Okada S, Tsumura M, Kagawa R, Miki M, Kawaguchi H, Nakamura K, Boisson-Dupuis S, Casanova JL, Takihara Y, Kobayashi M. Hirata O, et al. Haematologica. 2013 Oct;98(10):1641-9. doi: 10.3324/haematol.2013.083741. Epub 2013 Apr 12. Haematologica. 2013. PMID: 23585529 Free PMC article. - Biallelic TRAF3IP2 variants causing chronic mucocutaneous candidiasis in a child harboring a STAT1 variant.
Blanco Lobo P, Lei WT, Pelham SJ, Guisado Hernández P, Villaoslada I, de Felipe B, Lucena JM, Casanova JL, Olbrich P, Puel A, Neth O. Blanco Lobo P, et al. Pediatr Allergy Immunol. 2021 Nov;32(8):1804-1812. doi: 10.1111/pai.13603. Epub 2021 Aug 9. Pediatr Allergy Immunol. 2021. PMID: 34289170 Review. - The genetics of nontuberculous mycobacterial infection.
Newport M. Newport M. Expert Rev Mol Med. 2003 Feb 28;5(6):1-13. doi: 10.1017/S1462399403005908. Expert Rev Mol Med. 2003. PMID: 14987409 Review.
Cited by
- Naive and memory human B cells have distinct requirements for STAT3 activation to differentiate into antibody-secreting plasma cells.
Deenick EK, Avery DT, Chan A, Berglund LJ, Ives ML, Moens L, Stoddard JL, Bustamante J, Boisson-Dupuis S, Tsumura M, Kobayashi M, Arkwright PD, Averbuch D, Engelhard D, Roesler J, Peake J, Wong M, Adelstein S, Choo S, Smart JM, French MA, Fulcher DA, Cook MC, Picard C, Durandy A, Klein C, Holland SM, Uzel G, Casanova JL, Ma CS, Tangye SG. Deenick EK, et al. J Exp Med. 2013 Nov 18;210(12):2739-53. doi: 10.1084/jem.20130323. Epub 2013 Nov 11. J Exp Med. 2013. PMID: 24218138 Free PMC article. - Incomplete penetrance in primary immunodeficiency: a skeleton in the closet.
Gruber C, Bogunovic D. Gruber C, et al. Hum Genet. 2020 Jun;139(6-7):745-757. doi: 10.1007/s00439-020-02131-9. Epub 2020 Feb 17. Hum Genet. 2020. PMID: 32067110 Free PMC article. Review. - Inherited and acquired immunodeficiencies underlying tuberculosis in childhood.
Boisson-Dupuis S, Bustamante J, El-Baghdadi J, Camcioglu Y, Parvaneh N, El Azbaoui S, Agader A, Hassani A, El Hafidi N, Mrani NA, Jouhadi Z, Ailal F, Najib J, Reisli I, Zamani A, Yosunkaya S, Gulle-Girit S, Yildiran A, Cipe FE, Torun SH, Metin A, Atikan BY, Hatipoglu N, Aydogmus C, Kilic SS, Dogu F, Karaca N, Aksu G, Kutukculer N, Keser-Emiroglu M, Somer A, Tanir G, Aytekin C, Adimi P, Mahdaviani SA, Mamishi S, Bousfiha A, Sanal O, Mansouri D, Casanova JL, Abel L. Boisson-Dupuis S, et al. Immunol Rev. 2015 Mar;264(1):103-20. doi: 10.1111/imr.12272. Immunol Rev. 2015. PMID: 25703555 Free PMC article. Review. - A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.
Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. Michalska A, et al. Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review. - Inborn errors of immunity with loss- and gain-of-function germline mutations in STAT1.
Asano T, Utsumi T, Kagawa R, Karakawa S, Okada S. Asano T, et al. Clin Exp Immunol. 2023 Apr 25;212(2):96-106. doi: 10.1093/cei/uxac106. Clin Exp Immunol. 2023. PMID: 36420581 Free PMC article. Review.
References
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. - PubMed
- Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem. 2006;281:14111–8. - PubMed
- Averbuch D, Chapgier A, Boisson-Dupuis S, Casanova JL, Engelhard D. The clinical spectrum of patients with deficiency of Signal Transducer and Activator of Transcription1. Pediatr Infect Dis J. 2011;30:352–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN261200800001C/CA/NCI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- Z01 AI000647-16/ImNIH/Intramural NIH HHS/United States
- Z99 AI999999/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous