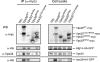Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy - PubMed (original) (raw)
Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy
Nobuo N Noda et al. J Biol Chem. 2012.
Abstract
Vacuolar protein sorting 30 (Vps30)/autophagy-related protein 6 (Atg6) is a common component of two distinct phosphatidylinositol 3-kinase complexes. In complex I, Atg14 links Vps30 to Vps34 lipid kinase and exerts its specific role in autophagy, whereas in complex II, Vps38 links Vps30 to Vps34 and plays a crucial role in vacuolar protein sorting. However, the molecular role of Vps30 in each pathway remains unclear. Here, we report the crystal structure of the carboxyl-terminal domain of Vps30. The structure is a novel globular fold comprised of three β-sheet-α-helix repeats. Truncation analyses showed that the domain is dispensable for the construction of both complexes, but is specifically required for autophagy through the targeting of complex I to the pre-autophagosomal structure. Thus, the domain is named the β-α repeated, autophagy-specific (BARA) domain. On the other hand, the N-terminal region of Vps30 was shown to be specifically required for vacuolar protein sorting. These structural and functional investigations of Vps30 domains, which are also conserved in the mammalian ortholog, Beclin 1, will form the basis for studying the molecular functions of this protein family in various biological processes.
Figures
FIGURE 1.
Structure of Vps30 BARA. A, overall structure of Vps30 BARA. The α-helices and β-strands are indicated with red helical ribbons and cyan arrows, respectively. Secondary structures are labeled, and residues adjacent to the disordered regions are numbered. Amino and carboxy termini are denoted as N and C, respectively. B, topology of Vps30 BARA. The α-helices and β-strands are indicated with boxes and arrows, respectively. Repeats 1, 2, and 3 are colored green, red, and cyan, respectively. The disordered regions are indicated with a broken line. C, superimposition of the secondary structural elements of repeats 2 and 3 on those of repeat 1. Coloring is as in B. D, left, ribbon representation of the three-helix bundle of BARA. Side chains involved in the interaction between helices are indicated with a stick model. Coloring is as in B. Right, ribbon representation of secondary structural elements of BARA. The side chains of conserved hydrophobic residues on the three sheets, which are bound to the grooves formed between helices, are indicated with a stick model. Coloring is as in B. All figures representing molecular structures were generated with PyMOL (47).
FIGURE 2.
Domain architecture of Vps30 and sequence alignment with homologues. A, domain architecture of Vps30. B, sequence alignment of the C-terminal region of Vps30/Beclin 1 homologues. Perfectly conserved residues are shaded black, and residues conserved as hydrophobic are shaded gray. The secondary structural elements of BARA are shown above the alignment. HsBec1, Homo sapiens Beclin 1; At, Arabidopsis thaliana; Dm, Drosophila melanogaster.
FIGURE 3.
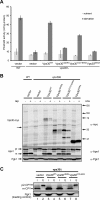
Assessment of autophagy and CPY-sorting activities. A, autophagic activity of _pho8_Δ_60 vps30_Δ yeast cells (TKY1308) expressing Vps30 mutants. Autophagic activity was measured using Pho8Δ60 alkaline phosphatase (ALP) assay as described under “Experimental Procedures.” Error bars indicate the standard deviation of three independent experiments. B, Ape1 maturation in _vps30_Δ cells expressing Vps30 mutants. Lysates of the _vps30_Δ cells (KVY135) carrying pRS314-based VPS30 mutants were subjected to Western blotting and Ape1 and Vps30 bands were detected using anti-Ape1 and anti-Myc antibodies, respectively. As a loading control, Pgk1 was detected using anti-Pgk1 antibody. C, CPY sorting in _vps30_Δ cells expressing Vps30 mutants. The _vps30_Δ cells (KVY135) carrying pRS314-based VPS30 mutants were subjected to CPY sorting assay as described under “Experimental Procedures.”
FIGURE 4.
Analysis of Vps30 interaction with Atg14 and Vps38. Coimmunoprecipitation experiments were performed as described under “Experimental Procedures.” Protein bands for Vps30, Atg14, Vps34, and Vps38 were detected using anti-Myc, anti-HA, anti-Vps34, and anti-HA antibodies, respectively. The samples used for the upper three panels were prepared from _vps30_Δ yeast cells (KVY135) expressing Vps30 mutants and Atg14-HA-GFP, whereas those used for the bottom panel were prepared from KVY135 cells expressing Vps30 mutants and Vps38-HA-GFP. Asterisks indicate degraded products of Vps30.
FIGURE 5.
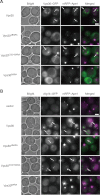
Vps30BARA is required for efficient PAS targeting of Vps30 and Atg14. A, the _vps30_Δ mRFP-APE1 yeast cells (TKY1647) expressing Vps30 mutants fused to GFP were subjected to microscopic observation. Vps30-GFP and mRFP-Ape1 in cells with rapamycin treatment for 1 h were observed using fluorescent microscopy. B, the _vps30_Δ mRFP-APE1 ATG14-GFP yeast cells (TKY1675) expressing Vps30 mutants were subjected to microscopic observation. Atg14-GFP and mRFP-Ape1 in cells with rapamycin treatment for 1 h were observed using fluorescent microscopy. Arrows indicate the PAS. A bar indicates 2 μm.
FIGURE 6.
Vps30CCD directly interacts with Atg14CCII. A, in vitro pulldown assay between GST-Atg14CCII and Vps30 mutants. Asterisks indicate degraded products of GST-Atg14CCII and Vps30CCD. B, purification of GST-Atg14CCII coexpressed with Vps30CCD in E. coli. After purification with GS4B beads (lane 1), GST was excised from Atg14CCII with PreScission protease (lane 2) and free GST was removed from the sample using GS4B beads (lane 3). The asterisk indicates a degradation product of Vps30CCD. Protein bands in A and B were stained with Coomassie Brilliant Blue.
FIGURE 7.
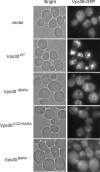
Vps30NTD is required for endosome-targeting of Vps38. The _VPS38-GFP vps30_Δ yeast cells (TKY1307) expressing Vps30 mutants in logarithmic phase were subjected to microscopic observation. Vps38-yEGFP was observed using fluorescent microscopy. A bar indicates 2 μm.
FIGURE 8.
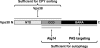
Summary of the functions of Vps30 domains revealed in this study.
Similar articles
- Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG.
Itakura E, Kishi C, Inoue K, Mizushima N. Itakura E, et al. Mol Biol Cell. 2008 Dec;19(12):5360-72. doi: 10.1091/mbc.e08-01-0080. Epub 2008 Oct 8. Mol Biol Cell. 2008. PMID: 18843052 Free PMC article. - Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Ohashi Y, Soler N, García Ortegón M, Zhang L, Kirsten ML, Perisic O, Masson GR, Burke JE, Jakobi AJ, Apostolakis AA, Johnson CM, Ohashi M, Ktistakis NT, Sachse C, Williams RL. Ohashi Y, et al. Autophagy. 2016 Nov;12(11):2129-2144. doi: 10.1080/15548627.2016.1226736. Epub 2016 Sep 14. Autophagy. 2016. PMID: 27630019 Free PMC article. - The Atg1 complex, Atg9, and Vac8 recruit PI3K complex I to the pre-autophagosomal structure.
Hitomi K, Kotani T, Noda NN, Kimura Y, Nakatogawa H. Hitomi K, et al. J Cell Biol. 2023 Aug 7;222(8):e202210017. doi: 10.1083/jcb.202210017. Epub 2023 Jul 12. J Cell Biol. 2023. PMID: 37436710 Free PMC article. - The class III phosphatidylinositol 3-kinase Vps34 in Saccharomyces cerevisiae.
Reidick C, Boutouja F, Platta HW. Reidick C, et al. Biol Chem. 2017 May 1;398(5-6):677-685. doi: 10.1515/hsz-2016-0288. Biol Chem. 2017. PMID: 27935849 Review. - Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Salminen A, et al. Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
Cited by
- Atomistic autophagy: the structures of cellular self-digestion.
Hurley JH, Schulman BA. Hurley JH, et al. Cell. 2014 Apr 10;157(2):300-311. doi: 10.1016/j.cell.2014.01.070. Cell. 2014. PMID: 24725401 Free PMC article. Review. - BECLIN 1-VPS34 COMPLEX ARCHITECTURE: UNDERSTANDING THE NUTS AND BOLTS OF THERAPEUTIC TARGETS.
Morris DH, Yip CK, Shi Y, Chait BT, Wang QJ. Morris DH, et al. Front Biol (Beijing). 2015 Oct;10(5):398-426. doi: 10.1007/s11515-015-1374-y. Epub 2015 Nov 4. Front Biol (Beijing). 2015. PMID: 26692106 Free PMC article. - BECN1s, a short splice variant of BECN1, functions in mitophagy.
Cheng B, Xu A, Qiao M, Wu Q, Wang W, Mei Y, Wu M. Cheng B, et al. Autophagy. 2015 Nov 2;11(11):2048-2056. doi: 10.1080/15548627.2015.1100785. Autophagy. 2015. PMID: 26649941 Free PMC article. - Conformational flexibility of BECN1: Essential to its key role in autophagy and beyond.
Mei Y, Glover K, Su M, Sinha SC. Mei Y, et al. Protein Sci. 2016 Oct;25(10):1767-85. doi: 10.1002/pro.2984. Epub 2016 Aug 13. Protein Sci. 2016. PMID: 27414988 Free PMC article. Review. - Biophysical characterization of Atg11, a scaffold protein essential for selective autophagy in yeast.
Suzuki H, Noda NN. Suzuki H, et al. FEBS Open Bio. 2017 Dec 4;8(1):110-116. doi: 10.1002/2211-5463.12355. eCollection 2018 Jan. FEBS Open Bio. 2017. PMID: 29321961 Free PMC article.
References
- Kametaka S., Okano T., Ohsumi M., Ohsumi Y. (1998) Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 273, 22284–22291 - PubMed
- Tsukada M., Ohsumi Y. (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 - PubMed
- Mizushima N., Yoshimori T., Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases