ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin - PubMed (original) (raw)
ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin
Daniel K Clare et al. Cell. 2012.
Abstract
The chaperonin GroEL assists the folding of nascent or stress-denatured polypeptides by actions of binding and encapsulation. ATP binding initiates a series of conformational changes triggering the association of the cochaperonin GroES, followed by further large movements that eject the substrate polypeptide from hydrophobic binding sites into a GroES-capped, hydrophilic folding chamber. We used cryo-electron microscopy, statistical analysis, and flexible fitting to resolve a set of distinct GroEL-ATP conformations that can be ordered into a trajectory of domain rotation and elevation. The initial conformations are likely to be the ones that capture polypeptide substrate. Then the binding domains extend radially to separate from each other but maintain their binding surfaces facing the cavity, potentially exerting mechanical force upon kinetically trapped, misfolded substrates. The extended conformation also provides a potential docking site for GroES, to trigger the final, 100° domain rotation constituting the "power stroke" that ejects substrate into the folding chamber.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
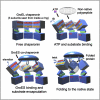
Diagram of the GroEL-GroES ATPase Cycle, Showing the Steps of Substrate Binding, Encapsulation, Folding, and Release ATP hydrolysis in one ring is required to enable subsequent ATP binding to the opposite ring, but hydrolysis is not required for folding to proceed within the chamber.
Figure 2
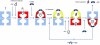
Cryo-EM Maps of the Seven Structures Determined from the GroEL-D398A-ATP Data Set Maps are shown as white surfaces. Top ring, side view, and bottom ring are shown for each complex. The atomic structures are fitted into the maps, with the equatorial domain in green, intermediate in yellow, and apical in red. T, tense allosteric state (unliganded); R, relaxed states (ATP bound). Rs rings are the ATP-bound rings from GroEL-ATP7 complexes (single), and Rd rings are from GroEL-ATP14 complexes (double). Figure produced with UCSF Chimera (Pettersen et al., 2004). See also Figures S1, S2, and S3.
Figure 3
Cut-Open Views of the EM Maps and Fits of Apo GroEL and the GroEL-ATP7 Complex with the Top Ring in the Rs1 State and the Bottom Ring in the T State The EM density is in white, and apical domain helices H and I, defining the substrate-binding site, are in red and orange, respectively. Intermediate domain helix M (green) contains the catalytic aspartate that contacts the nucleotide, and equatorial domain helix D (magenta) runs from the γ-phosphate to an inter-ring contact. The fitting shows that α-helical secondary structures are largely resolved in these maps. Shown below each complex is a view of the region around the ATP-binding pocket, which is empty in the T state but filled with density in the Rs1 state. The ATP molecule inside the Rs1 density is shown as spheres with CPK coloring. See also Figure S3.
Figure 4
Subunit Conformations and Intersubunit Salt Bridges in GroEL Rings Two adjacent subunits are shown as seen from inside the cavity, with EM density in white, helix H in red, helix I in orange, and helix M in green. Charged residues involved in intersubunit contacts are shown as spheres, with negatively charged residues in red, and positive ones in blue, and the contacts are listed for each ring. (A) GroEL-ATP7 structures. For comparison, the crystal conformation bound to GroES (PDB ID 1SVT) is also shown (R-ES). (B) GroEL-ATP14 structures. The arrows in the Rd series indicate the sequence of states inferred from the salt-bridge changes and the smallest movement in each step. The contact residues are listed in gray when the contact distance (defined in Table S2) is greater than 8 Å. See also Figures S4 and S5, Movies S1, S2, S3, and S4, and Tables S1 and S2.
Figure 5
Inter-ring Interfaces in the GroEL Complexes The salt-bridge contact involves E461 (red) and R452 (blue), along with the van der Waals contact V464-V464 (gray), seen in the left-hand contact in each panel. The contact at A109-A109 (gray, middle contact) is at the C-terminal end of helix D (magenta). D87 at the N terminus of helix D is involved in coordinating the ATP γ-phosphate. For the comparison with the GroES-bound state, we used the EM-derived model (PDB ID 2C7C) because the ring interface conformation in the crystal lattice is different from the one observed in solution (Ranson et al., 2006). The region viewed is indicated by the box in the overview (top, central panel). See also Figure S5, Movies S5, S6, S7, and S8, and Tables S1 and S2.
Figure 6
Footprint of GroES-Binding Sites on GroEL The rings are seen in space-filling format from above and in cut-away side view in (A) apo, (B) Rs1, (C) Rs-open, and (D) R-ES (PDB ID 1SVT) rings. Helices H and I are in red and orange, respectively, and the binding residues on the mobile loops of GroES are shown in dark blue. The black dotted circle shows the radial distribution of GroES-binding sites. In the R-open state, the sites are at the same radius as in R-ES, but they are rotated by ∼100°. GroES is schematically docked onto the R-open state to illustrate that the binding sites are readily accessible to the GroES mobile loops, unlike the situation in the Rs state. See also Figure S6.
Figure 7
Cartoon of Domain Movements with Helices H, I, and M Highlighted in Apo, Rs, R-open, and R-ES States Helices H (red), I (orange), and M (green) are highlighted. The folding substrate polypeptide is shown in gray mesh. GroES is shown docked onto the Rs-open state as in Figure 6C. The R-open apical domains have undergone about 70% of their elevation from apo to R-ES. This figure was generated in Blender 3D.
Figure S1
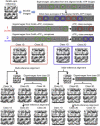
Schematic Outline of the Procedure Used to Sort the Data Set into Seven Distinct Structures, Related to Figure 2 The eigenimages of the whole, aligned data set indicate significant variations in the top and bottom of layers of the complexes, the locations of the apical domains. These eigenimages are circled in green. Class averages of this aligned data set show complexes with either one (red) or both (blue) apical domain layers extended relative to their appearance in apo GroEL. An initial rough sorting of the data into complexes with one (single, GroEL-ATP7) or both (double, GroEL-ATP14) rings occupied by ATP was based on the appearance of these classes. Eigenimages of the two subsets show more detailed features indicating variations in one or both apical domain layers. These eigenimages were used to further subdivide the two subsets, leading to the four structural classes whose 3D maps were used to generate reference projections for competitive alignment. Further eigenimage analysis led to one more subdivision of each set, yielding three single- and three double-ring ATP complexes.
Figure S2
C-Terminal Densities, Related to Figure 2 Cross-sections of the maps and fits showing the additional density arising from the GroEL C termini, which are disordered in the T state and in the crystal structures. They become more ordered in ATP-bound rings.
Figure S3
Flow Chart of Steps in the Flexible Fitting, Related to Figures 2 and 3 Lateral sheet superimposition in the equatorial domain contact was done by manual rigid fitting of the four strands from the 1OEL structure. All CG refinements were made without using map density. ATP was added by superimposing the equatorial domain from 1kp8. Mutations, rigid fitting, and segmentation done in Chimera, as were any manual changes.
Figure S4
Apical Domain Rotations, Related to Figure 4 The main apical domain movements are tabulated, and the orientations of a pair of adjacent apical domains in the representative conformations are shown with reference frames to indicate the view orientation and the axis and angle of rotation at each step.
Figure S5
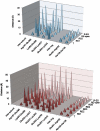
Plot of Intersubunit Contact Distances in the GroEL-ATP Complexes, Related to Figures 4 and 5 GroEL-ATP7 (top) and GroEL-ATP14 (bottom) complexes. The distances plotted are averaged over all seven subunits for each ring.
Figure S6
Surface Hydrophobicity Representations of Rings in the T, Rs1, Rs-open, and R-ES States, Related to Figure 6 The front half of each ring is cut away, with the cut surface shown in pale blue. Residue surfaces were colored using Chimera, based on the Kyte-Doolittle scale (Kyte and Doolittle, 1982), ranging from blue (most hydrophilic) through white to yellow (most hydrophobic). The continuous hydrophobic lining in the T state is distorted in Rs1, broken up in Rs-open, and occluded in R-ES.
Similar articles
- The C-terminal tails of the bacterial chaperonin GroEL stimulate protein folding by directly altering the conformation of a substrate protein.
Weaver J, Rye HS. Weaver J, et al. J Biol Chem. 2014 Aug 15;289(33):23219-23232. doi: 10.1074/jbc.M114.577205. Epub 2014 Jun 25. J Biol Chem. 2014. PMID: 24970895 Free PMC article. - Gly192 at hinge 2 site in the chaperonin GroEL plays a pivotal role in the dynamic apical domain movement that leads to GroES binding and efficient encapsulation of substrate proteins.
Machida K, Fujiwara R, Tanaka T, Sakane I, Hongo K, Mizobata T, Kawata Y. Machida K, et al. Biochim Biophys Acta. 2009 Sep;1794(9):1344-54. doi: 10.1016/j.bbapap.2008.12.003. Epub 2008 Dec 24. Biochim Biophys Acta. 2009. PMID: 19130907 - Effective ATPase activity and moderate chaperonin-cochaperonin interaction are important for the functional single-ring chaperonin system.
Illingworth M, Salisbury J, Li W, Lin D, Chen L. Illingworth M, et al. Biochem Biophys Res Commun. 2015 Oct 9;466(1):15-20. doi: 10.1016/j.bbrc.2015.08.034. Epub 2015 Aug 11. Biochem Biophys Res Commun. 2015. PMID: 26271593 - GroEL/GroES: structure and function of a two-stroke folding machine.
Xu Z, Sigler PB. Xu Z, et al. J Struct Biol. 1998 Dec 15;124(2-3):129-41. doi: 10.1006/jsbi.1998.4060. J Struct Biol. 1998. PMID: 10049801 Review. - GroEL and the GroEL-GroES Complex.
Ishii N. Ishii N. Subcell Biochem. 2017;83:483-504. doi: 10.1007/978-3-319-46503-6_17. Subcell Biochem. 2017. PMID: 28271487 Review.
Cited by
- Go hybrid: EM, crystallography, and beyond.
Lander GC, Saibil HR, Nogales E. Lander GC, et al. Curr Opin Struct Biol. 2012 Oct;22(5):627-35. doi: 10.1016/j.sbi.2012.07.006. Epub 2012 Jul 24. Curr Opin Struct Biol. 2012. PMID: 22835744 Free PMC article. Review. - Oligomerization of a molecular chaperone modulates its activity.
Saio T, Kawagoe S, Ishimori K, Kalodimos CG. Saio T, et al. Elife. 2018 May 1;7:e35731. doi: 10.7554/eLife.35731. Elife. 2018. PMID: 29714686 Free PMC article. - Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Liu C, Jin M, Wang S, Han W, Zhao Q, Wang Y, Xu C, Diao L, Yin Y, Peng C, Bao L, Wang Y, Cong Y. Liu C, et al. Commun Biol. 2023 May 16;6(1):531. doi: 10.1038/s42003-023-04915-x. Commun Biol. 2023. PMID: 37193829 Free PMC article. - The human mitochondrial Hsp60 in the APO conformation forms a stable tetradecameric complex.
Enriquez AS, Rojo HM, Bhatt JM, Molugu SK, Hildenbrand ZL, Bernal RA. Enriquez AS, et al. Cell Cycle. 2017 Jul 3;16(13):1309-1319. doi: 10.1080/15384101.2017.1321180. Epub 2017 Jun 8. Cell Cycle. 2017. PMID: 28594255 Free PMC article. - Crystal structure of a GroEL-ADP complex in the relaxed allosteric state at 2.7 Å resolution.
Fei X, Yang D, LaRonde-LeBlanc N, Lorimer GH. Fei X, et al. Proc Natl Acad Sci U S A. 2013 Aug 6;110(32):E2958-66. doi: 10.1073/pnas.1311996110. Epub 2013 Jul 16. Proc Natl Acad Sci U S A. 2013. PMID: 23861496 Free PMC article.
References
- Braig K., Otwinowski Z., Hegde R., Boisvert D.C., Joachimiak A., Horwich A.L., Sigler P.B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994;371:578–586. - PubMed
- Braig K., Adams P.D., Brünger A.T. Conformational variability in the refined structure of the chaperonin GroEL at 2.8 A resolution. Nat. Struct. Biol. 1995;2:1083–1094. - PubMed
- Burston S.G., Weissman J.S., Farr G.W., Fenton W.A., Horwich A.L. Release of both native and non-native proteins from a cis-only GroEL ternary complex. Nature. 1996;383:96–99. - PubMed
Supplemental Reference
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 070776/WT_/Wellcome Trust/United Kingdom
- 089050/WT_/Wellcome Trust/United Kingdom
- G0600084/MRC_/Medical Research Council/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- P41 RR017573/RR/NCRR NIH HHS/United States
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials