APOBEC3B and AID have similar nuclear import mechanisms - PubMed (original) (raw)
APOBEC3B and AID have similar nuclear import mechanisms
Lela Lackey et al. J Mol Biol. 2012.
Abstract
Members of the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) protein family catalyze DNA cytosine deamination and underpin a variety of immune defenses. For instance, several family members, including APOBEC3B (A3B), elicit strong retrotransposon and retrovirus restriction activities. However, unlike the other proteins, A3B is the only family member with steady-state nuclear localization. Here, we show that A3B nuclear import is an active process requiring at least one amino acid (Val54) within an N-terminal motif analogous to the nuclear localization determinant of the antibody gene diversification enzyme AID (activation-induced cytosine deaminase). Mechanistic conservation with AID is further suggested by A3B's capacity to interact with the same subset of importin proteins. Despite these mechanistic similarities, enforced A3B expression cannot substitute for AID-dependent antibody gene diversification by class switch recombination. Regulatory differences between A3B and AID are also visible during cell cycle progression. Our studies suggest that the present-day A3B enzyme retained the nuclear import mechanism of an ancestral AID protein during the expansion of the APOBEC3 locus in primates. Our studies also highlight the likelihood that, after nuclear import, specialized mechanisms exist to guide these enzymes to their respective physiological substrates and prevent gratuitous chromosomal DNA damage.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
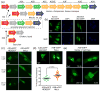
Figure 1. Relationships between AID and A3B
(a) Phylogenetic tree depicting the APOBEC loci in the indicated species. The split between fish and birds (~300 million years ago) and the divergence of the original placental mammal (~100 million years ago) are shown,,. (b) Representative images of HeLa cells transfected with human or zebrafish AID-eGFP and treated with lepB or ethanol as a vehicle control. (c) Representative images of HEK293T or HeLa cells expressing human AID-eGFP after treatment for the indicated times with lepB. (d) Representative images of HEK293T or HeLa cells expressing A3B-eGFP (quantified below; mean and SD shown for >20 individual cell measurements). (e) Representative images of A3B-eGFP expressed in the buccal tumor epithelial line TR146, the squamous cell carcinoma line JSQ3, the breast epithelial line MCF10A, and the osteosarcoma line U2OS.
Figure 2. A3B is actively imported into the nucleus
(a) Representative images of digitonin treated HeLa cells incubated with lysates containing GFP, A3B-eGFP, A3B NTD-eGFP, A3A-eGFP, or A3A NLS-eGFP. White arrows highlight instances of active nuclear import. (b) Quantification of the results from (a) using the same exposure for all conditions (FITC=1 second; n≥30 cells were examined and the mean nuclear fluorescent intensity is indicated for each condition). The inset is an anti-GFP immunoblot of representative cell lysates with asterisks indicating the correct bands.
Figure 3. A3B and AID localize differently during the mitosis
Representative image frames of (a) mCherry, (b) A3B-mCherry, and (c) AID-mCherry localization during HeLa cell cycle progression from late telophase of mitosis to early interphase. H2B-eGFP images of the same cells are shown below each time series. The white arrows at 0 min highlight informative cells, and the arrows at other time-points indicate significant localization events discussed in the text.
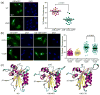
Figure 4. Single amino acid changes within the β2 region affects nuclear localization of A3B and AID
(a) Representative images of A3B-eGFP and A3B V54D-eGFP in HeLa cells. The adjacent dot plot reports quantification of the nuclear to total fluorescent signal (n≥25 cells were analyzed for each condition with mean and SD shown). (b) Representative images of AID-eGFP and AID Y48H-eGFP in HeLa cells taken 3 hrs after ethanol or lepB treatment. The adjacent dot plot reports quantification of the nuclear to total fluorescent signal (n≥50 cells were analyzed for each condition with mean and SD shown). Model ribbon structures of AID and A3B NTD depicted adjacent to the actual structure of A3G C-terminal domain (CTD). The β2 region and key amino acids in AID and A3B are labeled in red and circled; the side chains of zinc-coordinating residues are illustrated in green.
Figure 5. Both AID and A3B interact with members of the adaptor importin family
Immunoblots of input HEK293T protein lysates (bottom panel) and pulldown results for eGFP, AID-eGFP, AID Y48H-eGFP, A3B-eGFP and A3B V54D-eGFP with the indicated GST-importin substrates (top panels).
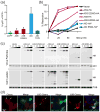
Figure 6. A3B does not perform class switch recombination and AID does not restrict HIV-1
(a) Flow cytometry quantification of the isotype switch to IgG in AID-deficient murine B cells expressing AID, A3B or non-functional mutants (mean and SD are shown for duplicate samples). (b) Infectivity of HIV-1 produced in HeLa cells expressing HA tagged APOBEC3G (A3G), A3B, AID and their catalytic mutants. High levels of restriction correspond to lower levels of fluorescence in a reporter CEM-GFP cell line and a decrease in infectious virus. Asterisks indicate that twice as much DNA was required for adequate expression of AID (0, 50, 100 and 200 ng). (c) Immunoblots of viral particle proteins (top) or cell lysate proteins (bottom) probed for anti-HA (APOBEC/AID expression), anti-p24 for a virus loading control, or anti-tubulin for a cellular loading control. (d) Representative images of mCherry, A3B-mCherry or AID-mCherry in HeLa cells 24 hrs after infection with replication competent HIV-1LAI nef::eGFP. Cells expressing the protein of interest infected with HIV are indicated by white arrows.
Similar articles
- APOBEC3B lysine residues are dispensable for DNA cytosine deamination, HIV-1 restriction, and nuclear localization.
Molan AM, Hanson HM, Chweya CM, Anderson BD, Starrett GJ, Richards CM, Harris RS. Molan AM, et al. Virology. 2017 Nov;511:74-81. doi: 10.1016/j.virol.2017.08.025. Epub 2017 Aug 23. Virology. 2017. PMID: 28841445 Free PMC article. - APOBEC3B Nuclear Localization Requires Two Distinct N-Terminal Domain Surfaces.
Salamango DJ, McCann JL, Demir Ö, Brown WL, Amaro RE, Harris RS. Salamango DJ, et al. J Mol Biol. 2018 Aug 17;430(17):2695-2708. doi: 10.1016/j.jmb.2018.04.044. Epub 2018 May 19. J Mol Biol. 2018. PMID: 29787764 Free PMC article. - The DNA deaminase APOBEC3B interacts with the cell-cycle protein CDK4 and disrupts CDK4-mediated nuclear import of Cyclin D1.
McCann JL, Klein MM, Leland EM, Law EK, Brown WL, Salamango DJ, Harris RS. McCann JL, et al. J Biol Chem. 2019 Aug 9;294(32):12099-12111. doi: 10.1074/jbc.RA119.008443. Epub 2019 Jun 19. J Biol Chem. 2019. PMID: 31217276 Free PMC article. - An overview of cytidine deaminases.
Navaratnam N, Sarwar R. Navaratnam N, et al. Int J Hematol. 2006 Apr;83(3):195-200. doi: 10.1532/IJH97.06032. Int J Hematol. 2006. PMID: 16720547 Review. - The AID/APOBEC family of nucleic acid mutators.
Conticello SG. Conticello SG. Genome Biol. 2008;9(6):229. doi: 10.1186/gb-2008-9-6-229. Epub 2008 Jun 17. Genome Biol. 2008. PMID: 18598372 Free PMC article. Review.
Cited by
- The preferred nucleotide contexts of the AID/APOBEC cytidine deaminases have differential effects when mutating retrotransposon and virus sequences compared to host genes.
Chen J, MacCarthy T. Chen J, et al. PLoS Comput Biol. 2017 Mar 31;13(3):e1005471. doi: 10.1371/journal.pcbi.1005471. eCollection 2017 Mar. PLoS Comput Biol. 2017. PMID: 28362825 Free PMC article. - APOBEC3B lysine residues are dispensable for DNA cytosine deamination, HIV-1 restriction, and nuclear localization.
Molan AM, Hanson HM, Chweya CM, Anderson BD, Starrett GJ, Richards CM, Harris RS. Molan AM, et al. Virology. 2017 Nov;511:74-81. doi: 10.1016/j.virol.2017.08.025. Epub 2017 Aug 23. Virology. 2017. PMID: 28841445 Free PMC article. - Ancestral APOBEC3B Nuclear Localization Is Maintained in Humans and Apes and Altered in Most Other Old World Primate Species.
Auerbach AA, Becker JT, Moraes SN, Moghadasi SA, Duda JM, Salamango DJ, Harris RS. Auerbach AA, et al. mSphere. 2022 Dec 21;7(6):e0045122. doi: 10.1128/msphere.00451-22. Epub 2022 Nov 14. mSphere. 2022. PMID: 36374108 Free PMC article. - DNA cytosine and methylcytosine deamination by APOBEC3B: enhancing methylcytosine deamination by engineering APOBEC3B.
Fu Y, Ito F, Zhang G, Fernandez B, Yang H, Chen XS. Fu Y, et al. Biochem J. 2015 Oct 1;471(1):25-35. doi: 10.1042/BJ20150382. Epub 2015 Jul 20. Biochem J. 2015. PMID: 26195824 Free PMC article. - D316 is critical for the enzymatic activity and HIV-1 restriction potential of human and rhesus APOBEC3B.
McDougle RM, Hultquist JF, Stabell AC, Sawyer SL, Harris RS. McDougle RM, et al. Virology. 2013 Jun 20;441(1):31-9. doi: 10.1016/j.virol.2013.03.003. Epub 2013 Mar 29. Virology. 2013. PMID: 23542011 Free PMC article.
References
- LaRue RS, Andrésdóttir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jónsson SR, Landau NR, Löchelt M, Malik HS, Malim MH, Münk C, O’Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–7. - PMC - PubMed
- Liao W, Hong SH, Chan BH, Rudolph FB, Clark SC, Chan L. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem Biophys Res Commun. 1999;260:398–404. - PubMed
- Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI083196/AI/NIAID NIH HHS/United States
- T32 AI007313/AI/NIAID NIH HHS/United States
- CAPMC/ CIHR/Canada
- P01 GM091743/GM/NIGMS NIH HHS/United States
- R01 AI064046/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources