Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation - PubMed (original) (raw)
Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation
Farzaneh Modarresi et al. Nat Biotechnol. 2012.
Abstract
The ability to specifically upregulate genes in vivo holds great therapeutic promise. Here we show that inhibition or degradation of natural antisense transcripts (NATs) by single-stranded oligonucleotides or siRNAs can transiently and reversibly upregulate locus-specific gene expression. Brain-derived neurotrophic factor (BDNF) is normally repressed by a conserved noncoding antisense RNA transcript, BDNF-AS. Inhibition of this transcript upregulates BDNF mRNA by two- to sevenfold, alters chromatin marks at the BDNF locus, leads to increased protein levels and induces neuronal outgrowth and differentiation both in vitro and in vivo. We also show that inhibition of NATs leads to increases in glial-derived neurotrophic factor (GDNF) and ephrin receptor B2 (EPHB2) mRNA. Our data suggest that pharmacological approaches targeting NATs can confer locus-specific gene upregulation effects.
Figures
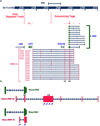
Figure 1. Genomic organization of the human BDNF locus showing
(A) genomic location of the BDNF sense and antisense transcripts and their relation to the other neighboring genes on chromosome 11. Solid boxes show exons and arrows show introns and direction of transcription. Different splice variants of _BDNF_-AS transcript are transcribed from the opposite DNA strand as that of BDNF mRNA. All _BDNF_-AS splice variants have a common exon that overlaps with 225 bp of all variants of BDNF mRNA. Inset data: Sequence tags generated by next-generation sequencing (RNA deep-seq), derived from human entorhinal cortex are aligned to the UCSC genome browser. Peaks represent nucleotide coverage, indicating reliable detection of _BDNF_-AS exons. (B) Identification of mouse _Bdnf_-AS by RACE experiments followed by sequencing. Proportional drawing representing human and mouse BDNF loci, showing direction of transcription for both BDNF and _BDNF_-AS transcripts, as well as the potential overlapping region. Mouse _Bdnf_-AS transcript is shorter than that of human and contains 1–2 exons. The potential overlapping region is bigger in mouse (934 bp compared to 225 bp in human), but the common overlap shows 90% homology across the two species. Real-time PCR (RT-PCR) primers, probes, and siRNAs are designed to target the non-overlapping parts of both transcripts. Blue Asterisks and numbers below each denote target sites of siRNAs used in this study, as well as m_Bdnf_-AntagoNAT3(*3) and m_Bdnf_-AntagoNAT9 (*9).
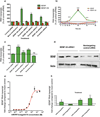
Figure 2. Antisense-mediated regulation of sense mRNA and protein
(A) Knockdown of brain derived neurotrophic factor (BDNF) natural antisense transcript, _BDNF_-AS, in HEK293T cells (n=12 per treatment) with each of three unique siRNAs (10 nM) targeting the non-overlapping region of _BDNF_-AS transcript, caused 2–6 fold upregulation of BDNF (sense) mRNA (n=6 for each data point/treatment ***= P < 0.001, **= P < 0.01). Similar results were obtained from experiments using Human cortical neuron (HCN), glioblastoma (MK059) cells, mouse N2a cells and neurospheres “data not shown”. Scrambled sequences, mock transfection and control siRNAs were used as controls. Control siRNA for this and other experiments is an inert siRNA (CCUCUCCACGCGCAGUACATT) that does not target any known sequence in the mammalian genome. All measurements were normalized to the 18S rRNA and graphed as a percentage of each mRNA to the negative siRNA control sample. (B) We assessed changes in BDNF and _BDNF_-AS transcripts over a period of time, following _BDNF_-AS knockdown (n=6 for each data point/treatment). siRNA knockdown of human _BDNF_-AS resulted in efficient and consistent downregulation of _BDNF_-AS, starting at 6 h and continuing on to 72 h. BDNF mRNA levels rose at 18 h, remaining high for more than 72 h, reversing to pre-treatment levels at 96 h. Note that the peak at 48 h is consistent and reproducible. Although _BDNF_-AS knockdown begins after 6 h, upregulation of BDNF started 18 h post-treatment. This time lag between the depletion of _BDNF_-AS and the increase of BDNF mRNA shows the sequential order of events indicating that the cells require time to adapt to the removal of the antisense transcript before upregulating BDNF. (C) siRNA-mediated knockdown of _BDNF_-AS transcript caused an increase in BDNF protein levels measured by ELISA. Cells were transfected with 10 nM of two active siRNAs for _BDNF_-AS, scrambled siRNAs or a control siRNA for 48 hours. The supernatants of these cells were concentrated and analyzed for BDNF protein by ELISA, using a commercially available kit. BDNF protein was significantly increased (n=6 per treatment, ***=P < 0.0001, **= P<0.001) with siRNA targeting _BDNF_-AS transcript. (D) Western blots confirmed that knockdown of the non-protein-coding _BDNF_-AS, with _BDNF_-AS siRNA1, but not control non-targeting siRNA transcript increased BDNF protein levels without changing the levels of beta-actin. Collectively, these data suggest that there is a discordant relationship between the sense and antisense BDNF transcripts in which _BDNF_-AS suppresses the expression of BDNF mRNA and protein. Removal of this negative regulatory effect, by _BDNF_-AS knockdown, causes upregulation of BDNF mRNA and protein levels. (E) Dose-dependent increases in Bdnf following _Bdnf_-AS depletion: We performed dose-response experiments using 11 different concentrations (1:3 serial dilutions ranging from 300nM to 5pM) of m_Bdnf_-AntagoNAT9 (n=6 per data point/treatment) and we observed a dose-dependent increase in Bdnf mRNA levels at 1–300 nM concentration with an EC50 of 6.6 nM. (F) Selective knockdown of GDNF-AS increases GDNF mRNA: We treated cells with various AntagoNATs targeting a low abundance noncoding antisense RNA, GDNF-AS. We observed that GDNF-AntagoNAT5 and GDNF-AntagoNAT6 increase the GDNF mRNA by 3–4 fold (n=6 per treatment *= P < 0.05, **= P < 0.01).
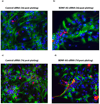
Figure 3. Bdnf upregulation increases neuronal outgrowth
(A–B) Immunocytochemistry images of hippocampal neurospheres treated with either control siRNA (A) or _Bdnf_-AS siRNA (B) 3 d post-platting. (C–D) Immunocytochemistry images of neuronal maturation and neurite outgrowth in hippocampal neurospheres treated with either control siRNA (C) or _Bdnf_-AS siRNA (D) 7 d post-plating. Treatment of cells with siRNA targeting the _Bdnf_-AS transcript resulted in increased neuronal cell number as well as increase in neurite outgrowth and maturation, both at 3d or 7d post-plating neurospheres. B-tubulin III stained red, GFAP stained green and DAPI stained blue.
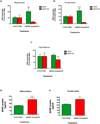
Figure 4. _Bdnf_-AS regulates Bdnf mRNA and protein in vivo
(A–C) Using osmotic mini-pumps, we infused m_Bdnf_-AntagoNAT9 (CAACATATCAGGAGCC) or control oligonucleotide (CCACGCGCAGTACATG) constantly over a period of 28 d, into the third ventricle of mouse brain (n=5 per treatment group *= P < 0.05, **= P < 0.01, ***= P < 0.001). m_Bdnf_-AntagoNAT9 directed against _Bdnf_-AS but not the control oligonucleotide resulted in an increase in Bdnf levels in the hippocampus (A) and frontal cortex (B). In the hypothalamus (C) both transcripts were unchanged, as was expected for a tissue that is not directly connected to the third ventricle of the brain. (D–E) We assessed BDNF protein levels by ELISA and found that m_Bdnf_-AntagoNAT9 treatment results in an increase in BDNF protein, both in the hippocampus (D) and frontal cortex (E), as compared to control oligonucleotide treated mice.
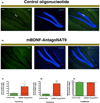
Figure 5. Blocking of _Bdnf_-AS, in vivo, causes an increase in neuronal survival and proliferation
(A–B) We treated mice with m_Bdnf_-AntagoNAT9 or control oligos. After 28 d of continuous m_Bdnf_-AntagoNAT9 infusion, we performed histological examination of brain tissues, using Ki67. Ki67 is the marker of proliferating cells in hippocampus and we observed an increase in the number of proliferating cells in mice received m_Bdnf_-AntagoNAT treatment compare to mice received control oligos. In mice treated with m_Bdnf_-AntagoNAT9 (B), there was an increase in Ki67 positive cells (proliferating cells), as compared to control treated mice (A). (C) Mice treated with m_Bdnf_-AntagoNAT9 had a significant increase in the number of Ki67 positive cells as compared to control treated mice. (D) In mice treated with m_Bdnf_-AntagoNAT9, there was a significant increase in the number of surviving cells (BrdU positive) as compared to control oligonucleotide treated mice. (E) There were no differences in hippocampal volume between control and m_Bdnf_-AntagoNAT9 treated mice. Together these data (n=5 per treatment group *= P < 0.05, ***= P < 0.001) demonstrates that _Bdnf_-AS regulates Bdnf levels in vivo and that blocking Bdnf sense-antisense interactions results in an increase in neuronal lineage, proliferation and survival.
Figure 6. Removal of _BDNF_-AS resulted in the modification of chromatin marks
(A) HEK293T cells were treated with control or _BDNF_-AS siRNA. 48 h post-transfection cells were harvested, fixed with formaldehyde, sonicated and incubated with antibodies against H3K27met3. Immunoprecipitation was performed followed by DNA extraction. DNA samples were analyzed using 16 primer sets covering the entire BDNF gene locus, and the BDNF promoter region, as indicated. (B) There was a decrease in association of the repressive chromatin marker, H3K27met3, upon treatment of the cells with _BDNF_-AS siRNA, both at the sense-antisense overlapping (primer-9) and promoter (primer12–14) regions (n=6 for each data point). The observed chromatin modification did not extend toward neighboring genes. These results suggest that local antisense-mediated chromatin modifications are occurring, beginning from the sense-antisense overlapping region and spreading in the 5’ direction towards the BDNF promoter region. (C) Knockdown of Ezh2, by either one of two different siRNAs, mimics or phenocopies the _BDNF_-AS knockdown and causes upregulation of BDNF mRNA (n=6 for each data point/treatment ***= P < 0.001). (D) ChIP assay using an Ezh2 antibody revealed that there was a decrease in Ezh2 association with the BDNF promoter upon depletion of the _BDNF_-AS transcript by siRNA. Not all 16 primer sets gave detectable PCR signals, possibly due to the lack of direct Ezh2-chromatin binding (n=6 for each data point/treatment **= P < 0.01, *= P < 0.05).
Comment in
- AntagoNATs boost gene expression.
Rusk N. Rusk N. Nat Methods. 2012 May;9(5):437. doi: 10.1038/nmeth.2007. Nat Methods. 2012. PMID: 22803202 No abstract available.
Similar articles
- What do natural antisense transcripts regulate?
Werner A, Carlile M, Swan D. Werner A, et al. RNA Biol. 2009 Jan-Mar;6(1):43-8. doi: 10.4161/rna.6.1.7568. Epub 2009 Jan 2. RNA Biol. 2009. PMID: 19098462 Review. - Inhibition of brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor expression reduces dopaminergic sprouting in the injured striatum.
Batchelor PE, Liberatore GT, Porritt MJ, Donnan GA, Howells DW. Batchelor PE, et al. Eur J Neurosci. 2000 Oct;12(10):3462-8. doi: 10.1046/j.1460-9568.2000.00239.x. Eur J Neurosci. 2000. PMID: 11029615 - Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis.
Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. Jin H, et al. BMC Mol Biol. 2008 Jan 14;9:6. doi: 10.1186/1471-2199-9-6. BMC Mol Biol. 2008. PMID: 18194570 Free PMC article. - Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters.
Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Pruunsild P, et al. Genomics. 2007 Sep;90(3):397-406. doi: 10.1016/j.ygeno.2007.05.004. Epub 2007 Jul 12. Genomics. 2007. PMID: 17629449 Free PMC article. - Long non-coding RNAs: From disease code to drug role.
Chen Y, Li Z, Chen X, Zhang S. Chen Y, et al. Acta Pharm Sin B. 2021 Feb;11(2):340-354. doi: 10.1016/j.apsb.2020.10.001. Epub 2020 Oct 10. Acta Pharm Sin B. 2021. PMID: 33643816 Free PMC article. Review.
Cited by
- GAS6-AS1 drives bladder cancer progression by increasing MMP7 expression in a ceRNA- and RBP-dependent manner.
Zhou X, Xiao L, Meng F, Zuo F, Wu W, Li G, Han F, Peng G, Shen H. Zhou X, et al. Transl Oncol. 2024 Oct;48:102065. doi: 10.1016/j.tranon.2024.102065. Epub 2024 Jul 24. Transl Oncol. 2024. PMID: 39053343 Free PMC article. - Role of the Long Non-Coding RNA MAPT-AS1 in Regulation of Microtubule Associated Protein Tau (MAPT) Expression in Parkinson's Disease.
Coupland KG, Kim WS, Halliday GM, Hallupp M, Dobson-Stone C, Kwok JB. Coupland KG, et al. PLoS One. 2016 Jun 23;11(6):e0157924. doi: 10.1371/journal.pone.0157924. eCollection 2016. PLoS One. 2016. PMID: 27336847 Free PMC article. - Non-coding RNAs as drug targets.
Matsui M, Corey DR. Matsui M, et al. Nat Rev Drug Discov. 2017 Mar;16(3):167-179. doi: 10.1038/nrd.2016.117. Epub 2016 Jul 22. Nat Rev Drug Discov. 2017. PMID: 27444227 Free PMC article. Review. - Small and Long Non-Coding RNAs: Novel Targets in Perspective Cancer Therapy.
Han Li C, Chen Y. Han Li C, et al. Curr Genomics. 2015 Oct;16(5):319-26. doi: 10.2174/1389202916666150707155851. Curr Genomics. 2015. PMID: 27047252 Free PMC article. - Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin-associated protein, HMGB2.
Yamanaka Y, Faghihi MA, Magistri M, Alvarez-Garcia O, Lotz M, Wahlestedt C. Yamanaka Y, et al. Cell Rep. 2015 May 12;11(6):967-976. doi: 10.1016/j.celrep.2015.04.011. Epub 2015 Apr 30. Cell Rep. 2015. PMID: 25937287 Free PMC article.
References
- Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. - PubMed
- Trzaska KA, et al. Brain-derived neurotrophic factor facilitates maturation of mesenchymal stem cell-derived dopamine progenitors to functional neurons. J Neurochem. 2009;110:1058–1069. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS063974-05/NS/NINDS NIH HHS/United States
- RC2 AG036596-01/AG/NIA NIH HHS/United States
- R01 NS063974-01A1/NS/NINDS NIH HHS/United States
- R01 NS063974/NS/NINDS NIH HHS/United States
- 5R01NS063974/NS/NINDS NIH HHS/United States
- RC2 AG036596-02/AG/NIA NIH HHS/United States
- 5RC2AG036596/AG/NIA NIH HHS/United States
- R01 NS063974-04/NS/NINDS NIH HHS/United States
- RC2 AG036596/AG/NIA NIH HHS/United States
- R01 NS063974-02/NS/NINDS NIH HHS/United States
- RC2 AG036596-03/AG/NIA NIH HHS/United States
- R01 NS063974-03/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous