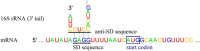Large variations in bacterial ribosomal RNA genes - PubMed (original) (raw)
Large variations in bacterial ribosomal RNA genes
Kyungtaek Lim et al. Mol Biol Evol. 2012 Oct.
Abstract
Ribosomal RNA (rRNA) genes, essential to all forms of life, have been viewed as highly conserved and evolutionarily stable, partly because very little is known about their natural variations. Here, we explored large-scale variations of rRNA genes through bioinformatic analyses of available complete bacterial genomic sequences with an emphasis on formation mechanisms and biological significance. Interestingly, we found bacterial genomes in which no 16S rRNA genes harbor the conserved core of the anti-Shine-Dalgarno sequence (5'-CCTCC-3'). This loss was accompanied by elimination of Shine-Dalgarno-like sequences upstream of their protein-coding genes. Those genomes belong to 1 or 2 of the following categories: primary symbionts, hemotropic Mycoplasma, and Flavobacteria. We also found many rearranged rRNA genes and reconstructed their history. Conjecturing the underlying mechanisms, such as inversion, partial duplication, transposon insertion, deletion, and substitution, we were able to infer their biological significance, such as co-orientation of rRNA transcription and chromosomal replication, lateral transfer of rRNA gene segments, and spread of rRNA genes with an apparent structural defect through gene conversion. These results open the way to understanding dynamic evolutionary changes of rRNA genes and the translational machinery.
Figures
Fig. 1.
Interaction between SD and anti-SD sequences. Interaction is between the mRNA 5′-UTR (for rpsB gene) and the 16S rRNA 3′ tail in Escherichia coli.
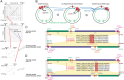
Fig. 2.
Comparative analysis of the anti-SD genomes and the non–anti-SD genomes in the class Flavobacteria. (A) Maximum likelihood phylogenetic tree. Groups 1 and 2 non-anti-SD genomes are shown in gray. (B) Predicted 16S rRNA 3′ tail sequences. The regions corresponding to the Escherichia coli anti-SD motif are shaded in gray. (C) SD indexes (d_F_SD). Triangle: cutoff value < −3.4535 kcal/mol; Dot: cutoff value < −4.4 kcal/mol. (D) Fractions of the four nucleotides (d_f_A, d_f_C, d_f_G, and d_f_T) at specific positions between −50 and −1 nt from all protein-coding genes in each genome. The background fraction was subtracted. For details, see Materials and Methods.
Fig. 3.
Comparative analysis of the anti-SD genomes and the non-anti-SD genomes in the phylum Proteobacteria. (A) Maximum likelihood phylogenetic tree. Group 3 non–anti-SD genomes are shown in gray. (B) Predicted 16S rRNA 3′ tail sequences. The regions corresponding to the Escherichia coli anti-SD motif are shaded in gray. (C) SD indexes (d_F_SD). Triangle: cutoff value < −3.4535 kcal/mol; Dot: cutoff value < −4.4 kcal/mol. (D) Fractions of the four nucleotides (d_f_A, d_f_C, d_f_G, and d_f_T) at specific positions between −50 and −1 nt from all protein-coding genes in each genome. The background fraction was subtracted. For details, see Materials and Methods. (i) The class Gammaproteobacteria. (ii) The class Betaproteobacteria. (iii) The class Alphaproteobacteria.
Fig. 4.
Comparative analysis of the anti-SD genomes and the non–anti-SD genomes in the genus Mycoplasma. (A) Maximum likelihood phylogenetic tree. Group 4 non–anti-SD genomes are shown in gray. (B) Predicted 16S rRNA 3′ tail sequences. The regions corresponding to the Escherichia coli anti-SD motif are shaded in gray. (C) SD indexes (d_F_SD). Triangle: cutoff value < −3.4535 kcal/mol; Dot: cutoff value < −4.4 kcal/mol. (D) Fractions of the four nucleotides (d_f_A, d_f_C, d_f_G, and d_f_T) at specific positions between −50 and −1 nt from all protein-coding genes in each genome. The background fraction was subtracted. For details, see Materials and Methods.
Fig. 5.
Inversion of rRNA operons. (A) Dotplot between genomes. (B) Reconstruction. (C) Recombination sites in one operon. (D) Recombination sites in the other operon. Amino acid names indicate the corresponding tRNA genes.
Fig. 6.
Partial duplication of rRNA genes. (A) Gene rearrangement in Actinobacillus pleuropneumoniae L20. (i) A proposed mechanism for the rearrangement: insertion with long target duplication. (ii) The other mechanism: tandem duplication followed by homologous recombination. (iii) The rearrangement. (B) Gene rearrangement in Bacillus thuringiensis BMB171. (i) A proposed mechanism for the rearrangement. (ii) The rearrangement.
Fig. 7.
An incomplete 16S rRNA gene on a plasmid. (A) Reconstruction. (B) The rearrangement. Two diverged regions between the two genes are boxed. (C) Sequence comparison in the diverged regions.
Fig. 8.
Transposon insertion. (A) An insertion into a 16S rRNA gene. (B) Another insertion into a 16S rRNA gene. Black arrows: target duplication. Gray arrows: incomplete inverted repeats at the transposon ends.
Fig. 9.
Other modes of rRNA gene rearrangements. (A) Internal deletion in a 16S rRNA gene. (B) Deletion in an rRNA operon. (C) Substitution. An amino acid name indicates the corresponding tRNA gene. (D) Inversion within an rRNA operon.
Similar articles
- Mycoplasma gallisepticum 16S rRNA genes.
Skamrov A, Goldman M, Klasova J, Beabealashvilli R. Skamrov A, et al. FEMS Microbiol Lett. 1995 May 15;128(3):321-5. doi: 10.1111/j.1574-6968.1995.tb07543.x. FEMS Microbiol Lett. 1995. PMID: 7781981 - Re-annotation of 12,495 prokaryotic 16S rRNA 3' ends and analysis of Shine-Dalgarno and anti-Shine-Dalgarno sequences.
Amin MR, Yurovsky A, Chen Y, Skiena S, Futcher B. Amin MR, et al. PLoS One. 2018 Aug 23;13(8):e0202767. doi: 10.1371/journal.pone.0202767. eCollection 2018. PLoS One. 2018. PMID: 30138483 Free PMC article. - [Speciation in bacteria: comparison of the 16S rRNA gene for closely related Enterococcus species].
Botina SG, Sukhodolets VV. Botina SG, et al. Genetika. 2006 Mar;42(3):325-30. Genetika. 2006. PMID: 16649658 Russian. - [16S rRNA gene sequence analysis for bacterial identification in the clinical laboratory].
Matsumoto T, Sugano M. Matsumoto T, et al. Rinsho Byori. 2013 Dec;61(12):1107-15. Rinsho Byori. 2013. PMID: 24605544 Review. Japanese. - Compilation and analysis of DNA sequences associated with apparent streptomycete promoters.
Strohl WR. Strohl WR. Nucleic Acids Res. 1992 Mar 11;20(5):961-74. doi: 10.1093/nar/20.5.961. Nucleic Acids Res. 1992. PMID: 1549509 Free PMC article. Review.
Cited by
- Simplification of Ribosomes in Bacteria with Tiny Genomes.
Nikolaeva DD, Gelfand MS, Garushyants SK. Nikolaeva DD, et al. Mol Biol Evol. 2021 Jan 4;38(1):58-66. doi: 10.1093/molbev/msaa184. Mol Biol Evol. 2021. PMID: 32681797 Free PMC article. - Structural basis of sequestration of the anti-Shine-Dalgarno sequence in the Bacteroidetes ribosome.
Jha V, Roy B, Jahagirdar D, McNutt ZA, Shatoff EA, Boleratz BL, Watkins DE, Bundschuh R, Basu K, Ortega J, Fredrick K. Jha V, et al. Nucleic Acids Res. 2021 Jan 11;49(1):547-567. doi: 10.1093/nar/gkaa1195. Nucleic Acids Res. 2021. PMID: 33330920 Free PMC article. - The Dynamic Interplay Between Ribosomal DNA and Transposable Elements: A Perspective From Genomics and Cytogenetics.
Garcia S, Kovarik A, Maiwald S, Mann L, Schmidt N, Pascual-Díaz JP, Vitales D, Weber B, Heitkam T. Garcia S, et al. Mol Biol Evol. 2024 Mar 1;41(3):msae025. doi: 10.1093/molbev/msae025. Mol Biol Evol. 2024. PMID: 38306580 Free PMC article. - Antimicrobial susceptibility and genetic profile of Mycoplasma hyopneumoniae isolates from Brazil.
Gonzaga NF, de Souza LFL, Santos MR, Assao VS, Rycroft A, Deeney AS, Fietto JLR, Bressan GC, Moreira MAS, Silva-Júnior A. Gonzaga NF, et al. Braz J Microbiol. 2020 Mar;51(1):377-384. doi: 10.1007/s42770-019-00185-0. Epub 2019 Dec 3. Braz J Microbiol. 2020. PMID: 31797326 Free PMC article. - Insights from 20 years of bacterial genome sequencing.
Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW. Land M, et al. Funct Integr Genomics. 2015 Mar;15(2):141-61. doi: 10.1007/s10142-015-0433-4. Epub 2015 Feb 27. Funct Integr Genomics. 2015. PMID: 25722247 Free PMC article. Review.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Chang B, Halgamuge S, Tang SL. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene. 2006;373:90–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources