Tetraspanin15 regulates cellular trafficking and activity of the ectodomain sheddase ADAM10 - PubMed (original) (raw)
Tetraspanin15 regulates cellular trafficking and activity of the ectodomain sheddase ADAM10
Johannes Prox et al. Cell Mol Life Sci. 2012 Sep.
Abstract
A disintegrin and metalloproteinase10 (ADAM10) has been implicated as a major sheddase responsible for the ectodomain shedding of a number of important surface molecules including the amyloid precursor protein and cadherins. Despite a well-documented role of ADAM10 in health and disease, little is known about the regulation of this protease. To address this issue we conducted a split-ubiquitin yeast two-hybrid screen to identify membrane proteins that interact with ADAM10. The yeast experiments and co-immunoprecipitation studies in mammalian cell lines revealed tetraspanin15 (TSPAN15) to specifically associate with ADAM10. Overexpression of TSPAN15 or RNAi-mediated knockdown of TSPAN15 led to significant changes in the maturation process and surface expression of ADAM10. Expression of an endoplasmic reticulum (ER) retention mutant of TSPAN15 demonstrated an interaction with ADAM10 already in the ER. Pulse-chase experiments confirmed that TSPAN15 accelerates the ER-exit of the ADAM10-TSPAN15 complex and stabilizes the active form of ADAM10 at the cell surface. Importantly, TSPAN15 also showed the ability to mediate the regulation of ADAM10 protease activity exemplified by an increased shedding of N-cadherin and the amyloid precursor protein. In conclusion, our data show that TSPAN15 is a central modulator of ADAM10-mediated ectodomain shedding. Therapeutic manipulation of its expression levels may be an additional approach to specifically regulate the activity of the amyloid precursor protein alpha-secretase ADAM10.
Figures
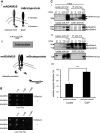
Fig. 1
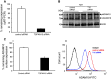
TSPAN15, a new ADAM10 interaction partner. a-I, II: Split-ubiquitin yeast two-hybrid system (modified from [50]). ADAM10 C-terminally fused to the C-terminal part of ubiquitin (Cub) and an artificial transcription factor (LexA-VP16) coexpressed in yeast together with a N-terminal NubG-tagged murine brain library. The close proximity between the ADAM10 bait protein and an interaction partner leads to the reconstitution of “split ubiquitin” to ubiquitin, which is recognized by cellular ubiquitin proteases, which in turn release the artificial transcription factor from the membrane. This enables yeast to grow on selective media [without leucine (-leu), tryptophane (-trp), histidine (-his)] plates. C-terminal part of ubiquitin (Cub, C), N-terminal part of ubiquitin (NubG, N), LexA (L). b ADAM10 bait protein coexpressed with the identified TSPAN15 prey protein and controls in NMY51 yeast. Transfection of both bait and prey protein is verified on Leu/Trp-lacking selective media plates, and interaction of ADAM10 and TSPAN15 is monitored under selective pressure on Leu/Trp/His-lacking media plates in comparison to controls (“+” and “−”). **c-**I: Mammalian expression constructs of murine ADAM10 and murine TSPAN15-myc were transiently coexpressed in HeLa cells. TSPAN15-myc (35–37 kDa) was precipitated using an anti-myc antibody, and coprecipitation of ADAM10 was detected with an anti-ADAM10 antibody. II: ADAM10 and TSPAN15-myc were transiently coexpressed in HeLa cells, and ADAM10 was precipitated using an anti-ADAM10 antibody. Coprecipitation was analyzed by Western blot using an anti-myc antibody. Single transfections of TSPAN15-myc and ADAM10 served as specificity controls for the antibodies used for immunoprecipitation. III: Quantification of band intensities of pro and mature form of ADAM10 in lysates and CoIP fractions. Ratios of band intensities of pro/(pro + mature) forms of ADAM10 were calculated (%, n = 5). Student’s t test was performed (***p < 0.005). [pro(p) ADAM10, 95 kDa, mature(m) ADAM10, 75 kDa]. Abbreviations: untransfected (Ø), vector (mock) transfected (V), murine ADAM10 (A10), murine TSPAN15-myc (T15), asterisk marks immunoglobulin signals
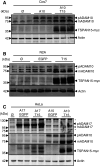
Fig. 2
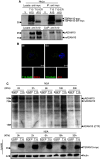
TSPAN15-myc influences ADAM10 maturation. a Murine ADAM10 transiently coexpressed with either EGFP or murine TSPAN15-myc in Cos7 cells. After immunoblotting ADAM10 was detected using an ADAM10 specific C-terminal antibody (B42.1) and TSPAN15-myc was detected using an anti-myc antibody. b TSPAN15-myc or EGFP was transiently expressed in N2A cells. After lysis proteins were immunoblotted, and endogenous ADAM10 and TSPAN15-myc were detected. c Murine ADAM17 or murine ADAM10 was transiently expressed either with EGFP or TSPAN15-myc in HeLa cells. Presence of ADAM17 [pro(p) ADAM17, closed arrowhead, 120 kDa; putative mature ADAM17, _open arrowhead_] was analyzed using a C-terminal-specific antibody, and TSPAN15-myc and ADAM10 expressions were monitored as mentioned above. Actin served as protein loading control. Asterisks mark unspecific antibody binding
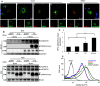
Fig. 3
TSPAN15-myc influences ADAM10 localization. **a-**I, II: Murine ADAM10 and ADAM10/TSPAN15-myc expressed in Cos7 cells. III, IV: Murine ADAM17 and ADAM17/TSPAN15-myc expressed in Cos7 cells. Confocal immunofluorescence pictures were taken using an anti-KDEL antibody, an anti-ADAM10 antibody, and an anti-ADAM17 antibody, respectively. Scale bar 100 μm. V, VI: SHSY cells were transiently transfected with a C-terminal EGFP-tagged variant of human TSPAN15. Endogenous ADAM10 was stained using an anti-ADAM10 antibody (11G2), and an anti-PDI antibody was used as ER marker. Asterisks mark untransfected cells. Scale bar 10 μm. b N2A cells were biotinylated after transfection with either EGFP or murine TSPAN15-myc. Following cell lysis total protein samples were taken, and after precipitation of biotin-labeled proteins immunoblotting of total lysates and bound fractions was performed. TSPAN15-myc and ADAM10 were detected. The detection of the transferrin receptor (TFR) was included as a control for biotinylated surface proteins, and antibodies against the intracellular glycerin-aldehyd-3-phosphate dehydrogenase (GAPDH) were used as a negative control. c-I: N2A cells were transfected with ADAM10, TSPAN15-myc and ADAM10/TSPAN15-myc; ADAM10 cell surface expression was determined through FACS analysis using an N-terminal anti-ADAM10 antibody. RFUs were determined. Statistical significance was determined using Student’s t test; values are provided as highly significant (**p < 0.01). Abbreviation: relative fluorescence units (RFU). II: Representative overlay of the FACS analysis performed
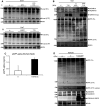
Fig. 4
Activity analysis of ADAM10. a-I: murine (m)ADAM10 expressed or coexpressed with TSPAN15-myc in Cos7 cells. Activity of ADAM10 was studied through the analysis of ADAM10-specific shedding events using immunoblot detection of N-cadherin [full-length (Fl) N-cadherin (130 kDa), C-terminal fragment (CTF) N-cadherin (37 kDa)] applying a C-terminal-specific N-cadherin antibody. II: EGFP and TSPAN15-myc were transiently expressed in Cos7 cells, and the processing of N-cadherin was analyzed by Western blot. b N2A cells were transfected with pcDNA3.1 or TSPAN15-myc and APP/pcDNA3.1 or APP/TSPAN15-myc. Processing of APP was analyzed using a C-terminal-specific anti-APP antibody. Actin served as protein loading control. c Cell culture supernatants of N2A cells transfected with EGFP or TSPAN15-myc and sAPPalpha content were determined using sandwich ELISAs. d HEK293 cells were transfected with human TSPAN15-EGFP or vector control, and endogenous APP processing was analyzed. APP (FL) APP full-length protein; APP (CTF) APP C-terminal C83 fragment; overex overexposed image of the upper panel. ADAM10, TSPAN15 and actin expression analysis was included
Fig. 5
Knockdown of TSPAN15. a Knockdown of TSPAN15 in N2A cells was verified through qRT-PCR analysis. b N2A cells were transfected with siRNA against TSPAN15 and control siRNA. After lysis and immunoblot analysis, ADAM10 was detected. Actin served as protein-loading control. c-I: Surface expression of ADAM10 was analyzed through FACS analysis using a N-terminal anti ADAM10 antibody. Remaining surface expression of ADAM10 was determined (%). II: Representative overlay of FACS analysis of ADAM10 surface protein after TSPAN15 knockdown
Fig. 6
Early interaction of TSPAN15 with ADAM10. a Murine ADAM10 was either expressed with TSPAN15-myc or TSPAN15-ER-myc. Cells were lysed, and myc-tagged proteins were precipitated using an anti-myc antibody. After immunoblotting coprecipitated ADAM10 was detected using a C-terminal-specific antibody (B42.1). **b-**I, II: Cos7 cells were transfected with TSPAN15-ER-myc or TSPAN15-ER-myc/ADAM10. Confocal immunofluorescence pictures were taken using an anti-myc antibody, an anti-KDEL antibody and an N-terminal-specific ADAM10 antibody and adequate secondary antibody pairs. Scale bar 100 μm. c N2A cells were transfected with EGFP or TSPAN15-myc, pulsed for 1 h with 35S methionine/cysteine and chased for 0, 2, 6, 18 and 30 h. After cell lysis equal amounts of protein were used, and ADAM10 was precipitated using a C-terminal-specific anti-ADAM10 antibody, subjected to SDS-PAGE and analyzed by fluorographics. TSPAN15-myc expression was analyzed by Western blot, and actin served as protein-loading control for the lysates
Fig. 7
Model of TSPAN15 function. (1) ADAM10 is synthesized as an inactive precursor (pro-ADAM10) in the ER, and TSPAN15 accelerates its ER exit. (2) After ER exit the immature ADAM10 is activated through removal of the inhibitory prodomain by furin or proprotein convertase (PC7), and is then transported to the plasma membrane together with TSPAN15. (3) At the plasma membrane TSPAN15 facilitates the integration of ADAM10 in the tetraspanin web. (4, 5) ADAM10 is stabilized, and the web composition enables ADAM10 to get access to its substrates and subsequent cleavage events in the juxtamembrane regions of the substrates take place. (6) Afterwards remaining membrane-bound fragments are cleaved through ripping proteases (γ-secretase complex/SPPLs) in the transmembrane regions, thereby liberating soluble intracellular domains (ICD), which might have signaling functions
Similar articles
- TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals.
Dornier E, Coumailleau F, Ottavi JF, Moretti J, Boucheix C, Mauduit P, Schweisguth F, Rubinstein E. Dornier E, et al. J Cell Biol. 2012 Oct 29;199(3):481-96. doi: 10.1083/jcb.201201133. Epub 2012 Oct 22. J Cell Biol. 2012. PMID: 23091066 Free PMC article. - TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization.
Jouannet S, Saint-Pol J, Fernandez L, Nguyen V, Charrin S, Boucheix C, Brou C, Milhiet PE, Rubinstein E. Jouannet S, et al. Cell Mol Life Sci. 2016 May;73(9):1895-915. doi: 10.1007/s00018-015-2111-z. Epub 2015 Dec 19. Cell Mol Life Sci. 2016. PMID: 26686862 Free PMC article. - Tetraspanin 3: A central endocytic membrane component regulating the expression of ADAM10, presenilin and the amyloid precursor protein.
Seipold L, Damme M, Prox J, Rabe B, Kasparek P, Sedlacek R, Altmeppen H, Willem M, Boland B, Glatzel M, Saftig P. Seipold L, et al. Biochim Biophys Acta Mol Cell Res. 2017 Jan;1864(1):217-230. doi: 10.1016/j.bbamcr.2016.11.003. Epub 2016 Nov 3. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 27818272 - Regulation of the trafficking and the function of the metalloprotease ADAM10 by tetraspanins.
Saint-Pol J, Eschenbrenner E, Dornier E, Boucheix C, Charrin S, Rubinstein E. Saint-Pol J, et al. Biochem Soc Trans. 2017 Aug 15;45(4):937-44. doi: 10.1042/BST20160296. Epub 2017 Jul 7. Biochem Soc Trans. 2017. PMID: 28687716 Review. - Regulation of ADAM10 by the TspanC8 Family of Tetraspanins and Their Therapeutic Potential.
Harrison N, Koo CZ, Tomlinson MG. Harrison N, et al. Int J Mol Sci. 2021 Jun 23;22(13):6707. doi: 10.3390/ijms22136707. Int J Mol Sci. 2021. PMID: 34201472 Free PMC article. Review.
Cited by
- Tetraspanin 15 depletion impairs extracellular vesicle docking at target neurons.
Stajano D, Lombino FL, Schweizer M, Glatzel M, Saftig P, Gromova KV, Kneussel M. Stajano D, et al. J Extracell Biol. 2023 Sep 11;2(9):e113. doi: 10.1002/jex2.113. eCollection 2023 Sep. J Extracell Biol. 2023. PMID: 38938373 Free PMC article. - GW501516-Mediated Targeting of Tetraspanin 15 Regulates ADAM10-Dependent N-Cadherin Cleavage in Invasive Bladder Cancer Cells.
Barbaud A, Lascombe I, Péchery A, Arslan S, Kleinclauss F, Fauconnet S. Barbaud A, et al. Cells. 2024 Apr 19;13(8):708. doi: 10.3390/cells13080708. Cells. 2024. PMID: 38667323 Free PMC article. - Dynamic association of the intramembrane proteases SPPL2a/b and their substrates with tetraspanin-enriched microdomains.
Leinung N, Mentrup T, Patel M, Gallagher T, Schröder B. Leinung N, et al. iScience. 2023 Sep 4;26(10):107819. doi: 10.1016/j.isci.2023.107819. eCollection 2023 Oct 20. iScience. 2023. PMID: 37736044 Free PMC article. - Structural basis for membrane-proximal proteolysis of substrates by ADAM10.
Lipper CH, Egan ED, Gabriel KH, Blacklow SC. Lipper CH, et al. Cell. 2023 Aug 17;186(17):3632-3641.e10. doi: 10.1016/j.cell.2023.06.026. Epub 2023 Jul 28. Cell. 2023. PMID: 37516108 Free PMC article. - The versatile roles of ADAM8 in cancer cell migration, mechanics, and extracellular matrix remodeling.
Mierke CT. Mierke CT. Front Cell Dev Biol. 2023 Feb 23;11:1130823. doi: 10.3389/fcell.2023.1130823. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36910158 Free PMC article. Review.
References
- Lopez-Perez E, Zhang Y, Frank SJ, Creemers J, Seidah N, Checler F. Constitutive alpha-secretase cleavage of the beta-amyloid precursor protein in the furin-deficient LoVo cell line: involvement of the pro-hormone convertase 7 and the disintegrin metalloprotease ADAM10. J Neurochem. 2001;76(5):1532–1539. doi: 10.1046/j.1471-4159.2001.00180.x. - DOI - PubMed
- Lopez-Perez E, Seidah NG, Checler F. Proprotein convertase activity contributes to the processing of the Alzheimer’s beta-amyloid precursor protein in human cells: evidence for a role of the prohormone convertase PC7 in the constitutive alpha-secretase pathway. J Neurochem. 1999;73(5):2056–2062. - PubMed
- Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15(10):1837–1839. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous