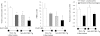Resolvin D1 receptor stereoselectivity and regulation of inflammation and proresolving microRNAs - PubMed (original) (raw)
Resolvin D1 receptor stereoselectivity and regulation of inflammation and proresolving microRNAs
Sriram Krishnamoorthy et al. Am J Pathol. 2012 May.
Abstract
Resolution of acute inflammation is an active process that involves the biosynthesis of specialized proresolving lipid mediators. Among them, resolvin D1 (RvD1) actions are mediated by two G protein-coupled receptors (GPCRs), ALX/FPR2 and GPR32, that also regulate specific microRNAs (miRNAs) and their target genes in novel resolution circuits. We report the ligand selectivity of RvD1 activation of ALX/FPR2 and GPR32. In addition to RvD1, its aspirin-triggered epimer and RvD1 analogs each dose dependently and effectively activated ALX/FPR2 and GPR32 in GPCR-overexpressing β-arrestin systems using luminescence and electric cell-substrate impedance sensing. To corroborate these findings in vivo, neutrophil infiltration in self-limited peritonitis was reduced in human ALX/FPR2-overexpressing transgenic mice that was further limited to 50% by RvD1 treatment with as little as 10 ng of RvD1 per mouse. Analysis of miRNA expression revealed that RvD1 administration significantly up-regulated miR-208a and miR-219 in exudates isolated from ALX/FPR2 transgenic mice compared with littermates. Overexpression of miR-208a in human macrophages up-regulated IL-10. In comparison, in ALX/FPR2 knockout mice, RvD1 neither significantly reduced leukocyte infiltration in zymosan-induced peritonitis nor regulated miR-208a and IL-10 in these mice. Together, these results demonstrate the selectivity of RvD1 interactions with receptors ALX/FPR2 and GPR32. Moreover, they establish a new molecular circuit that is operative in the resolution of acute inflammation activated by the proresolving mediator RvD1 involving specific GPCRs and miRNAs.
Copyright © 2012 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures
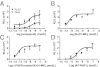
Figure 1
RvD1, AT-RvD1, and their stable analogs directly act on hALX/FPR2. Ligand-receptor interaction was monitored using the HEK-ALX/FPR2 β-arrestin system (see Materials and Methods). Dose-response activation curves were obtained for RvD1 and DHA (A), RvD1-ME (B), 17(R/S)-methyl RvD1-ME (C), and AT-RvD1 (D). Results are expressed as mean ± SEM (n = 3 to 4). RLU, relative luminescence units.
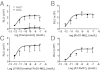
Figure 2
RvD1, AT-RvD1, and their stable analogs directly act on hGPR32. Ligand-receptor interactions were determined using the GPR32 β-arrestin system as in Figure 1. Dose-response activation curves are shown for RvD1 and DHA (A), RvD1-ME (B), 17(R/S)-methyl RvD1-ME (C), and AT-RvD1 (D). Results are expressed as mean ± SEM (n = 4 to 6). RLU, relative luminescence units.
Figure 3
RvD1, Rv analogs, and compound 43 produce hGPR32-dependent changes in impedance. Ligand-receptor–dependent changes in impedance were continuously recorded with real-time monitoring across cell monolayers using an ECIS system with hGPR32-expressing β-arrestin cells. Results are tracings obtained from incubations with CHO-GPR32 β-arrestin cells plus RvD1 (1, 10, and 100 nmol/L) (A); compound 43 (1, 10, and 100 nmol/L) (B); 100 nmol/L RvD1, RvD1-ME, and 17(R/S)-methyl RvD1-ME (C); and 100 nmol/L RvD1 directly compared with native DHA (D). Each tracing is representative of n = 3. Insets in A and B show dose-dependent changes in cell impedance determined at 5 minutes of incubation of GPR32-overexpressing cells with RvD1 or compound 43 or vehicle alone. Results are expressed as mean ± SEM (n = 3).
Figure 4
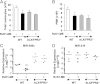
RvD1 reduction in PMN infiltration is enhanced in hALX/FPR2-overexpressing transgenic (Tg) mice peritonitis. hALX/FPR2-Tg mice and WT littermates were injected with zymosan (1 mg per mouse i.p.), with or without RvD1-ME at 10 ng per mouse i.v. Inflammatory exudates were collected at 24 hours and total exudate leukocytes were determined (see Materials and Methods): total leukocytes (left panel) and PMNs (middle panel). The right panel shows numbers of total leukocytes and resident macrophages in control mice. Results are expressed as mean ± SEM (n = 3 mice). *P < 0.05, WT, zymosan versus zymosan + RvD1; †P = 0.05, zymosan, WT versus ALX/FPR2-Tg; ‡P < 0.01, zymosan + RvD1 WT versus ALX/FPR2-Tg.
Figure 5
RvD1 stimulates specific proresolving miRNAs in hALX/FPR2 transgenic (Tg) mice. miRNA fractions were isolated from peritonitis exudate samples of WT littermates and ALX/FPR2-Tg mice challenged with zymosan (1 mg per mouse) compared with mice given RvD1 (10 ng per mouse with zymosan). Real-time PCR analyses of indicated miRNAs were performed and results were analyzed by the 2−ΔCT method. Fold change in expression levels in RvD1-treated mice compared with zymosan alone were calculated from 2−ΔCT values. Results are expressed as mean ± SEM (n = 3 in each group) fold change in levels of miR-208a (A), miR-219 (B), and miR-21, miR-146b, and miR-302d (C). *P < 0.05 for RvD1-treated versus zymosan-treated WT or Tg mice; †P < 0.05 for zymosan + RvD1–treated Tg mice versus zymosan + RvD1–treated WT mice. D: miR-208a and miR-219 expression levels determined by real-time PCR in peritoneal cells from untreated mice.
Figure 6
Regulation of acute inflammation and miRNA by RvD1 in ALX/FPR2 knockout mice. Number of total leukocytes (A) and PMNs (B) in peritoneal lavage fluid from WT or ALX/FPR2−/− mice injected with zymosan (1 mg per mouse, i.p.) with or without RvD1-ME (10 ng per mouse, i.v.). Lavage fluid samples were collected 24 hours after initiation of peritonitis, and cells were stained with anti-Ly-6G and F4/80 antibodies. Results are expressed as mean ± SEM (n = 6 mice per group). *P < 0.05 versus zymosan-treated group. Expression of miR-208a (C) and miR-219 (D) in exudate cells from WT or ALX/FPR2−/− mice 24 hours after injection of zymosan plus RvD1-ME (10 ng per mouse, i.v.) or zymosan plus saline. Relative expression of miRNAs was determined using real-time PCR. Results are expressed as mean ± SEM of fold changes in expression (n = 6 mice per group). **P < 0.001 versus zymosan-treated group.
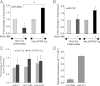
Figure 7
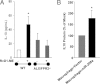
Regulation of IL-10 levels by RvD1 in peritonitis and by miR-208a in human macrophages. A: IL-10 levels were determined in mALX/FPR2−/− mice (n = 3 to 6). *P < 0.05 versus mice receiving zymosan alone. B: IL-10 levels were determined in human macrophages transiently overexpressing miR-208a or control vector (n = 3). *P < 0.05 versus control vector–transfected macrophages.
Similar articles
- MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits.
Recchiuti A, Krishnamoorthy S, Fredman G, Chiang N, Serhan CN. Recchiuti A, et al. FASEB J. 2011 Feb;25(2):544-60. doi: 10.1096/fj.10-169599. Epub 2010 Oct 18. FASEB J. 2011. PMID: 20956612 Free PMC article. - Resolvin D1 limits polymorphonuclear leukocyte recruitment to inflammatory loci: receptor-dependent actions.
Norling LV, Dalli J, Flower RJ, Serhan CN, Perretti M. Norling LV, et al. Arterioscler Thromb Vasc Biol. 2012 Aug;32(8):1970-8. doi: 10.1161/ATVBAHA.112.249508. Epub 2012 Apr 12. Arterioscler Thromb Vasc Biol. 2012. PMID: 22499990 Free PMC article. - The resolvin D1 receptor GPR32 transduces inflammation resolution and atheroprotection.
Arnardottir H, Thul S, Pawelzik SC, Karadimou G, Artiach G, Gallina AL, Mysdotter V, Carracedo M, Tarnawski L, Caravaca AS, Baumgartner R, Ketelhuth DF, Olofsson PS, Paulsson-Berne G, Hansson GK, Bäck M. Arnardottir H, et al. J Clin Invest. 2021 Dec 15;131(24):e142883. doi: 10.1172/JCI142883. J Clin Invest. 2021. PMID: 34699386 Free PMC article. - Resolvin D1 and its GPCRs in resolution circuits of inflammation.
Recchiuti A. Recchiuti A. Prostaglandins Other Lipid Mediat. 2013 Dec;107:64-76. doi: 10.1016/j.prostaglandins.2013.02.004. Epub 2013 Mar 15. Prostaglandins Other Lipid Mediat. 2013. PMID: 23500003 Review. - The lipoxin receptor ALX: potent ligand-specific and stereoselective actions in vivo.
Chiang N, Serhan CN, Dahlén SE, Drazen JM, Hay DW, Rovati GE, Shimizu T, Yokomizo T, Brink C. Chiang N, et al. Pharmacol Rev. 2006 Sep;58(3):463-87. doi: 10.1124/pr.58.3.4. Pharmacol Rev. 2006. PMID: 16968948 Review.
Cited by
- The resolution code of acute inflammation: Novel pro-resolving lipid mediators in resolution.
Serhan CN, Chiang N, Dalli J. Serhan CN, et al. Semin Immunol. 2015 May;27(3):200-15. doi: 10.1016/j.smim.2015.03.004. Epub 2015 Apr 7. Semin Immunol. 2015. PMID: 25857211 Free PMC article. Review. - CCL2 Inhibition of Pro-Resolving Mediators Potentiates Neuroinflammation in Astrocytes.
Gutiérrez IL, Novellino F, Caso JR, García-Bueno B, Leza JC, Madrigal JLM. Gutiérrez IL, et al. Int J Mol Sci. 2022 Mar 18;23(6):3307. doi: 10.3390/ijms23063307. Int J Mol Sci. 2022. PMID: 35328727 Free PMC article. - Immune responsive resolvin D1 programs myocardial infarction-induced cardiorenal syndrome in heart failure.
Halade GV, Kain V, Serhan CN. Halade GV, et al. FASEB J. 2018 Jul;32(7):3717-3729. doi: 10.1096/fj.201701173RR. Epub 2018 Feb 13. FASEB J. 2018. PMID: 29455574 Free PMC article. - The Anti-Inflammatory Mediator 17(R)-Resolvin D1 Attenuates Pressure Overload-Induced Cardiac Hypertrophy and Fibrosis.
Wang M, Pan W, Wei C, Liu J, Zhang J, Yu J, Zhao M, Xu S, Ye J, Wang Z, Ye D, Feng Y, Xu Y, Wan J. Wang M, et al. Drug Des Devel Ther. 2023 Oct 11;17:3073-3083. doi: 10.2147/DDDT.S421894. eCollection 2023. Drug Des Devel Ther. 2023. PMID: 37849783 Free PMC article. - Resolvins: Endogenously-Generated Potent Painkilling Substances and their Therapeutic Perspectives.
Yoo S, Lim JY, Hwang SW. Yoo S, et al. Curr Neuropharmacol. 2013 Dec;11(6):664-76. doi: 10.2174/1570159X11311060009. Curr Neuropharmacol. 2013. PMID: 24396341 Free PMC article.
References
- Brink C., Dahlén S.-E., Drazen J., Evans J.F., Hay D.W.P., Nicosia S., Serhan C.N., Shimizu T., Yokomizo T. International Union of Pharmacology XXXVII: nomenclature for leukotriene and lipoxin receptors. Pharmacol Rev. 2003;55:195–227. - PubMed
- Shimizu T. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu Rev Pharmacol Toxicol. 2009;49:123–150. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases