Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops - PubMed (original) (raw)
. 2012 Apr 10;109(15):5663-8.
doi: 10.1073/pnas.1112391109. Epub 2012 Mar 26.
Andrés Finzi, Xueling Wu, Cajetan Dogo-Isonagie, Lawrence K Lee, Lucas R Moore, Stephen D Schmidt, Jonathan Stuckey, Yongping Yang, Tongqing Zhou, Jiang Zhu, David A Vicic, Asim K Debnath, Lawrence Shapiro, Carole A Bewley, John R Mascola, Joseph G Sodroski, Peter D Kwong
Affiliations
- PMID: 22451932
- PMCID: PMC3326499
- DOI: 10.1073/pnas.1112391109
Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops
Young Do Kwon et al. Proc Natl Acad Sci U S A. 2012.
Abstract
The HIV-1 envelope (Env) spike (gp120(3)/gp41(3)) undergoes considerable structural rearrangements to mediate virus entry into cells and to evade the host immune response. Engagement of CD4, the primary human receptor, fixes a particular conformation and primes Env for entry. The CD4-bound state, however, is prone to spontaneous inactivation and susceptible to antibody neutralization. How does unliganded HIV-1 maintain CD4-binding capacity and regulate transitions to the CD4-bound state? To define this mechanistically, we determined crystal structures of unliganded core gp120 from HIV-1 clades B, C, and E. Notably, all of these unliganded HIV-1 structures resembled the CD4-bound state. Conformational fixation with ligand selection and thermodynamic analysis of full-length and core gp120 interactions revealed that the tendency of HIV-1 gp120 to adopt the CD4-bound conformation was restrained by the V1/V2- and V3-variable loops. In parallel, we determined the structure of core gp120 in complex with the small molecule, NBD-556, which specifically recognizes the CD4-bound conformation of gp120. Neutralization by NBD-556 indicated that Env spikes on primary isolates rarely assume the CD4-bound conformation spontaneously, although they could do so when quaternary restraints were loosened. Together, the results suggest that the CD4-bound conformation represents a "ground state" for the gp120 core, with variable loop and quaternary interactions restraining unliganded gp120 from "snapping" into this conformation. A mechanism of control involving deformations in unliganded structure from a functionally critical state (e.g., the CD4-bound state) provides advantages in terms of HIV-1 Env structural diversity and resistance to antibodies and inhibitors, while maintaining elements essential for entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Unliganded structures of HIV-1 gp120 core. Crystal structures are displayed as Cα-ribbon diagrams with outer domains in gray and inner domains in magenta, blue or light blue, cyan, and green for HIV-1 clades B, C, and E, and SIV, respectively, and the region that in the CD4-bound conformation makes up the bridging sheet in red. The evolutionary relationships of these Env glycoproteins are represented by a dendrogram where the length of connections is proportional to evolutionary distance. [The four HIV-1 structures were determined here; the SIV structure was determined previously by Chen et al. (17).]
Fig. 2.
Comparison of unliganded HIV-1 gp120 core to previously determined gp120 structures. Despite substantial conformational diversity of the gp120 envelope glycoprotein, unliganded HIV-1 gp120 snaps into a conformation that closely resembles the CD4-bound state. (A) Ribbon diagrams, displayed as in Fig. 1. (B) Molecular surface representation colored by structural deviations from the HIV-1 clade E unliganded gp120 structure. The color scale ranges from dark blue to red for rmsds of 0 to >10 Å, respectively. Notably, conformational changes >100 Å are observed in the bridging sheet region between the b13-bound and unliganded forms of HIV-1 gp120.
Fig. 3.
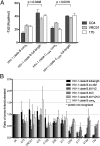
Conformational diversity of HIV-1 gp120 in solution. The conformational diversity of gp120 in solution is sensitive to the presence of the variable loops. (A) Entropy of ligand interactions with full-length and truncated gp120s (for measurements involving the HIV-1 clade B gp120, the YU2 strain was used; for measurements involving the HIV-1 clade C gp120, the C1086 strain was used). (B) Conformational fixation followed by ligand selection. The ratio of ligand binding to cross-linked vs. untreated gp120s is shown for full-length and truncated forms of gp120. ps, recognition by pooled sera from HIV-1-infected individuals, which was used for normalization.
Fig. 4.
Conformational diversity of HIV-1 gp120 as assessed by the small molecule NBD-556 and mechanism of control. The unliganded state of HIV-1 gp120 exists as an equilibrium of conformations, with the coree portion of gp120 displaying a strong intrinsic propensity to snap into the CD4-bound conformation when not restrained by variable loops or by interactions with gp41. (A) Structure of small molecule NBD-556 in complex with HIV-1 gp120 provides atomic-level details of its binding site, and also provides an explanation for its preference for the CD4-bound conformation of gp120. The surface of coree gp120 is colored blue for inner domain, gray for outer domain, and red for bridging sheet, with the small molecule NBD-556 binding at a highly conserved pocket at the nexus of inner domain, outer domain, and bridging sheet minidomain. The NBD-556 is shown in stick representation, colored yellow for carbon, red for oxygen, blue for nitrogen, and green for chlorine. (B) Close-up rotated 90° about a vertical axis from A. (C) Assessment of gp120 conformation in solution. SPR measurements of NBD-556 binding to gp120 in coree (Left) or full-length (Right) gp120 contexts and by direct binding (Upper) or by competition (Lower) indicates that the unliganded coree has a greater propensity to assume the CD4-bound conformation than full-length gp120. (D) Assessment of gp120 conformation in the functional viral spike. Neutralization by NBD-556 or NBD-557 is strongly enhanced by gp41 mutants (Upper) and to a lesser extent by changes in the V1/V2 region (Lower).
Fig. 5.
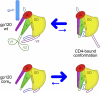
Mechanism of gp120 conformation control. In full-length WT gp120 (Upper), interactions with gp41 and between the variable loops V1/V2 and V3 shift the conformational equilibrium of the gp120 core away from the CD4-bound conformation (equilibrium is denoted by size of blue arrows). The intrinsic propensity of coree gp120 (Lower) to assume the CD4-bound conformation is revealed when the gp41-interactive region and variable loops are removed.
Similar articles
- Residues in the gp41 Ectodomain Regulate HIV-1 Envelope Glycoprotein Conformational Transitions Induced by gp120-Directed Inhibitors.
Pacheco B, Alsahafi N, Debbeche O, Prévost J, Ding S, Chapleau JP, Herschhorn A, Madani N, Princiotto A, Melillo B, Gu C, Zeng X, Mao Y, Smith AB 3rd, Sodroski J, Finzi A. Pacheco B, et al. J Virol. 2017 Feb 14;91(5):e02219-16. doi: 10.1128/JVI.02219-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28003492 Free PMC article. - Range of CD4-Bound Conformations of HIV-1 gp120, as Defined Using Conditional CD4-Induced Antibodies.
Kaplan G, Roitburd-Berman A, Lewis GK, Gershoni JM. Kaplan G, et al. J Virol. 2016 Apr 14;90(9):4481-4493. doi: 10.1128/JVI.03206-15. Print 2016 May. J Virol. 2016. PMID: 26889042 Free PMC article. - Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.
Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. Wang H, et al. Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):E7151-E7158. doi: 10.1073/pnas.1615939113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799557 Free PMC article. - Structure-based design, synthesis and validation of CD4-mimetic small molecule inhibitors of HIV-1 entry: conversion of a viral entry agonist to an antagonist.
Courter JR, Madani N, Sodroski J, Schön A, Freire E, Kwong PD, Hendrickson WA, Chaiken IM, LaLonde JM, Smith AB 3rd. Courter JR, et al. Acc Chem Res. 2014 Apr 15;47(4):1228-37. doi: 10.1021/ar4002735. Epub 2014 Feb 6. Acc Chem Res. 2014. PMID: 24502450 Free PMC article. Review. - Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.
Liu Q, Zhang P, Lusso P. Liu Q, et al. Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
Cited by
- Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies.
Henderson R, Anasti K, Manne K, Stalls V, Saunders C, Bililign Y, Williams A, Bubphamala P, Montani M, Kachhap S, Li J, Jaing C, Newman A, Cain DW, Lu X, Venkatayogi S, Berry M, Wagh K, Korber B, Saunders KO, Tian M, Alt F, Wiehe K, Acharya P, Alam SM, Haynes BF. Henderson R, et al. Nat Commun. 2024 Nov 3;15(1):9503. doi: 10.1038/s41467-024-53120-9. Nat Commun. 2024. PMID: 39489734 Free PMC article. - CD4 downregulation precedes Env expression and protects HIV-1-infected cells from ADCC mediated by non-neutralizing antibodies.
Richard J, Sannier G, Zhu L, Prévost J, Marchitto L, Benlarbi M, Beaudoin-Bussières G, Kim H, Sun Y, Chatterjee D, Medjahed H, Bourassa C, Delgado G-G, Dubé M, Kirchhoff F, Hahn BH, Kumar P, Kaufmann DE, Finzi A. Richard J, et al. mBio. 2024 Nov 13;15(11):e0182724. doi: 10.1128/mbio.01827-24. Epub 2024 Oct 7. mBio. 2024. PMID: 39373535 Free PMC article. - The asymmetric opening of HIV-1 Env by a potent CD4 mimetic enables anti-coreceptor binding site antibodies to mediate ADCC.
Richard J, Grunst MW, Niu L, Díaz-Salinas MA, Tolbert WD, Marchitto L, Zhou F, Bourassa C, Yang D, Chiu TJ, Chen HC, Benlarbi M; Guillaume-Beaudoin-Buissières; Gottumukkala S, Li W, Dionne K, Bélanger É, Chatterjee D, Medjahed H, Hendrickson WA, Sodroski J, Lang ZC, Morton AJ, Huang RK, Matthies D, Smith AB 3rd, Mothes W, Munro JB, Pazgier M, Finzi A. Richard J, et al. bioRxiv [Preprint]. 2024 Aug 27:2024.08.27.609961. doi: 10.1101/2024.08.27.609961. bioRxiv. 2024. PMID: 39253431 Free PMC article. Preprint. - ADS-J21 is a novel HIV-1 entry inhibitor targeting gp41.
Liang R, Dou D, Wang C, Huo S, Wu Y, Wang J, Yu Z, Zhang S, Xu J, Liu Y, Liu P, Jiang S, Yu F. Liang R, et al. Curr Res Microb Sci. 2024 Jul 9;7:100260. doi: 10.1016/j.crmicr.2024.100260. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39129758 Free PMC article. - Antiviral Protein-Protein Interaction Inhibitors.
Marković V, Szczepańska A, Berlicki Ł. Marković V, et al. J Med Chem. 2024 Mar 14;67(5):3205-3231. doi: 10.1021/acs.jmedchem.3c01543. Epub 2024 Feb 23. J Med Chem. 2024. PMID: 38394369 Free PMC article. Review.
References
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. - PubMed
- Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. - PubMed
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. - PubMed
- Sattentau QJ. HIV gp120: Double lock strategy foils host defences. Structure. 1998;6:945–949. - PubMed
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials