Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans - PubMed (original) (raw)
Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans
Cynthia Portal-Celhay et al. BMC Microbiol. 2012.
Abstract
Background: A powerful approach to understanding complex processes such as aging is to use model organisms amenable to genetic manipulation, and to seek relevant phenotypes to measure. Caenorhabditis elegans is particularly suited to studies of aging, since numerous single-gene mutations have been identified that affect its lifespan; it possesses an innate immune system employing evolutionarily conserved signaling pathways affecting longevity. As worms age, bacteria accumulate in the intestinal tract. However, quantitative relationships between worm genotype, lifespan, and intestinal lumen bacterial load have not been examined. We hypothesized that gut immunity is less efficient in older animals, leading to enhanced bacterial accumulation, reducing longevity. To address this question, we evaluated the ability of worms to control bacterial accumulation as a functional marker of intestinal immunity.
Results: We show that as adult worms age, several C. elegans genotypes show diminished capacity to control intestinal bacterial accumulation. We provide evidence that intestinal bacterial load, regulated by gut immunity, is an important causative factor of lifespan determination; the effects are specified by bacterial strain, worm genotype, and biologic age, all acting in concert.
Conclusions: In total, these studies focus attention on the worm intestine as a locus that influences longevity in the presence of an accumulating bacterial population. Further studies defining the interplay between bacterial species and host immunity in C. elegans may provide insights into the general mechanisms of aging and age-related diseases.
Figures
Figure 1
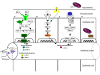
Signaling pathways important for C. elegans intestinal defenses against bacterial proliferation. A. DAF-2 insulin/IGF-I like signaling pathway. Activation of the DAF-2 receptor results in the phosphorylation of the phosphatidyl inositol 3 kinase (AGE-1) which catalyses the conversion of phosphatidylinositol biphosphate (PiP2) into phosphatidylinositol triphosphate (PiP3). The kinases PDK-1 and AKT-1/AKT-2 are activated by PiP3, which inhibits the transcription factor DAF-16. Relief of this inhibition leads to the expression of a set of stress response and antimicrobial genes. B. p38 MAPK pathway. PMK-1 is homologous to the mammalian p38 MAPK and acts downstream of NSY-1/MAKK kinase kinase and SEK-1/MAPK kinase. No interaction between TOL-1 and TIR-1 has been demonstrated. C. TGF-β pathway. The TGF-β homologue DBL-1 binds to the heterodimeric receptor SMA-6/DAF-4 and signals through the Smad proteins SMA-2, SMA-3 and SMA-4, which activate the transcription of genes involved in regulation of body size and innate immunity. The expression of lysozyme gene lys-1 is under the control of the p38 MAPK pathway and the DBL-1/TGF-β pathway. D. Mitochondrial enzymes. CLK-1 is an enzyme required for the biosynthesis of ubiquinoe CoQ9, an acceptor of electrons from both complexes I and II in C. elegans cells. Decreased complex I-dependent respiration of clk-1 mutants leads to decreased ROS production with lengthening lifespan and slowing development. TRX-1 is a mitochondrial oxidoreductase with important roles in lifespan regulation and oxidative stress response.
Figure 2
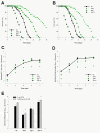
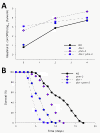
Density of bacterial accumulation in the C. elegans intestine by worm age and genotype, and bacterial strain. Survival of N2 C. elegans and DAF-2 pathway mutants when grown on lawns of E. coli OP50 (Panel A) or S. typhimurium SL1344 (Panel B). Intestinal density of viable E. coli OP50 (Panel C) or S. typhimurium SL1344 (Panel D) in N2 C. elegans and DAF-2 pathway mutants. Panel E: Intestinal load of E. coli OP50 (dark bars) or S. typhimurium SL1344 (grey bars) within N2 C. elegans and DAF-2 pathway mutants on day 2 (L4 stage + 2 days) of their lifespan. Data represent Mean ± SD from experiments involving 30 worms/group. Significant difference (p < 0.05) compared to N2 worms exposed to E. coli OP50 or S. typhimurium SL1344, indicated by * or **, respectively.
Figure 3
daf-16 mutation partially suppresses the daf-2 bacterial proliferation phenotypes in C. elegans. Panel A: Survival of daf-2, daf-16 single mutants, and daf-16;daf-2 double mutant when grown on lawns of E. coli OP50. Panel B: Intestinal density of viable E. coli OP50 in the intestine of the single and daf-16;daf-2 double mutants.
Figure 4
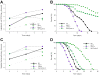
Survival and density of colonizing bacteria in the intestine of C. elegans mutants with altered immune function. Panel A: Survival of N2 C. elegans and four mutants with altered intestinal immune function when grown on lawns of E. coli OP50. Panel B: Intestinal load of E. coli OP50 (dark bars) or S. typhimurium SL1344 (grey bars) within N2 C. elegans and the four mutants with altered intestinal immune function on day 2 (L4 stage + 2 days) of their lifespan. Data represent Mean ± SD from experiments involving 30 worms/group. Significant differences (p < 0.05) compared to N2 worms exposed to E. coli OP50 or S. typhimurium SL1344, indicated by * or **, respectively. Panel C: Survival of daf-2 and dbl-1 single mutants, and the daf-2;dbl-1 double mutant when grown on lawns of E. coli OP50. Panel D: Intestinal density of viable E. coli OP50 in the intestine of the single and daf-2;dbl-1 double mutants. The dbl-1 mutation suppresses both the daf-2 intestinal bacterial proliferation and lifespan phenotypes.
Figure 5
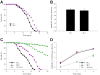
Role of downstream components of the innate immunity pathways on intestinal bacterial proliferation and C. elegans lifespan. Survival of C. elegans mutants with defective expression of antimicrobial peptides (Panel A) or oxidative stress enzymes (Panel C) when grown on lawns of E. coli OP50. Panel B: Intestinal load of E. coli OP50 (dark bars) or S. typhimurium SL1344 (grey bars) with altered intestinal expression of antimicrobial peptides or oxidative stress enzymes (Panel D) on day 2 (L4 stage + 2 days) of their lifespan. Data represent Mean ± SD from experiments involving 30 worms/group. Significant (p < 0.05) differences in proliferation either E. coli or Salmonella compared to N2 worms indicated by *.
Figure 6
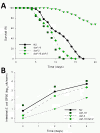
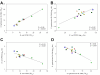
Relationship between C. elegans genotype, colonizing bacterial species, and lifespan. Symbols for the 14 worm genotypes are as indicated in Table 1. Panel A: Relationship of lifespans for worms grown on E. coli OP50 and S. typhimurium SL1344, measured as TD50. Worm survival is strongly correlated with growth on the two organisms (R = 0.98; p < 0.0001). Panel B: Relationship of intestinal bacterial density for worms grown on E. coli OP50 or S. typhimurium, measured as day 2 log10 cfu. Results show a strong direct correlation for the two bacterial species (R = 0.82; p < 0.001). Panel C: Relationship between lifespan and intestinal bacterial density for C. elegans grown on E. coli OP50 lawns. There is an inverse correlation between intestinal bacterial density and survival (R = 0.83; p < 0.001). Panel D: Relationship between lifespan and intestinal bacterial density for C. elegans grown on S. typhimurium SL1344 lawns. There is an inverse correlation between intestinal bacterial density and survival (R = 0.89; p < 0.001).
Figure 7
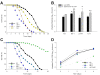
Density of colonizing bacteria in the intestine of C. elegans strains and their survival. Number (cfu) of E. coli OP50 (Panel A) or S. typhimurium SL1344 (Panel C) within the intestine of N2 (wild type), daf-2 and phm-2 single mutant, and daf-2;phm-2 double mutant C. elegans strains**. Panel B)** Survival of same strains when grown on lawns of E. coli OP50 or S. typhimurium SL1344 (Panel D).
Figure 8
C. elegans daf-2 mutants do not require a functional grinder to control intestinal bacterial proliferation. Fluorescence microscopy of N2, daf-2 and phm-2 single mutant, and daf-2;phm-2 double mutant C. elegans strains feeding on GFP-expressing E. coli.
Figure 9
Immunocompromised C. elegans are hypersusceptible to bacterial accumulation. Panel A: Number (cfu) of E. coli OP50 within the intestine of N2, dbl-1 and phm-2 single mutant, and dbl-1;phm-2 double mutant C. elegans strains**. Panel B:** Survival of same strains when grown on lawns of E. coli OP50.
Figure 10
Daf-2 mutation suppresses the clk-1 mitochondrion-dependent intestinal bacterial proliferation phenotype. Survival of N2 C. elegans and clk-1 mutants when grown on lawns of E. coli OP50 (Panel A). Panel B: Intestinal load of E. coli OP50 within N2 C. elegans and clk-1 mutants on day 2 (L4 stage + 2 days) of their lifespan. Data represent Mean ± SD from experiments involving 30 worms/group. Panel C: Survival of daf-2 and clk-1 single mutants and the daf-2;clk-1 double mutant when grown on lawns of E. coli OP50. Panel D: Intestinal density of viable E. coli OP50 in the intestine of the daf-2 and clk-1 single mutants and the daf-2;clk-1 double mutants.
Similar articles
- Lifespan Extension in C. elegans Caused by Bacterial Colonization of the Intestine and Subsequent Activation of an Innate Immune Response.
Kumar S, Egan BM, Kocsisova Z, Schneider DL, Murphy JT, Diwan A, Kornfeld K. Kumar S, et al. Dev Cell. 2019 Apr 8;49(1):100-117.e6. doi: 10.1016/j.devcel.2019.03.010. Dev Cell. 2019. PMID: 30965033 Free PMC article. - Probiotic Lactobacillus fermentum strain JDFM216 stimulates the longevity and immune response of Caenorhabditis elegans through a nuclear hormone receptor.
Park MR, Ryu S, Maburutse BE, Oh NS, Kim SH, Oh S, Jeong SY, Jeong DY, Oh S, Kim Y. Park MR, et al. Sci Rep. 2018 May 10;8(1):7441. doi: 10.1038/s41598-018-25333-8. Sci Rep. 2018. PMID: 29748542 Free PMC article. - Bacteria and the aging and longevity of Caenorhabditis elegans.
Kim DH. Kim DH. Annu Rev Genet. 2013;47:233-46. doi: 10.1146/annurev-genet-111212-133352. Annu Rev Genet. 2013. PMID: 24274752 Review. - [C. elegans defence mechanisms].
Ziegler K, Pujol N. Ziegler K, et al. Med Sci (Paris). 2009 May;25(5):497-503. doi: 10.1051/medsci/2009255497. Med Sci (Paris). 2009. PMID: 19480831 Review. French.
Cited by
- Quiescence Enhances Survival during Viral Infection in Caenorhabditis elegans.
Iannacone MJ, Um P, Grubbs JI, van der Linden AM, Raizen DM. Iannacone MJ, et al. J Neurosci. 2024 Aug 28;44(35):e1700222024. doi: 10.1523/JNEUROSCI.1700-22.2024. J Neurosci. 2024. PMID: 39060176 - Pathogen-induced Caenorhabditis elegans developmental plasticity has a hormetic effect on the resistance to biotic and abiotic stresses.
Leroy M, Mosser T, Manière X, Alvarez DF, Matic I. Leroy M, et al. BMC Evol Biol. 2012 Sep 21;12:187. doi: 10.1186/1471-2148-12-187. BMC Evol Biol. 2012. PMID: 22998555 Free PMC article. - 5-Dodecanolide interferes with biofilm formation and reduces the virulence of Methicillin-resistant Staphylococcus aureus (MRSA) through up regulation of agr system.
Valliammai A, Sethupathy S, Priya A, Selvaraj A, Bhaskar JP, Krishnan V, Pandian SK. Valliammai A, et al. Sci Rep. 2019 Sep 24;9(1):13744. doi: 10.1038/s41598-019-50207-y. Sci Rep. 2019. PMID: 31551455 Free PMC article. - Neural control of behavioral and molecular defenses in C. elegans.
Singh J, Aballay A. Singh J, et al. Curr Opin Neurobiol. 2020 Jun;62:34-40. doi: 10.1016/j.conb.2019.10.012. Epub 2019 Dec 5. Curr Opin Neurobiol. 2020. PMID: 31812835 Free PMC article. Review. - Using Bacterial Transcriptomics to Investigate Targets of Host-Bacterial Interactions in Caenorhabditis elegans.
Chan JP, Wright JR, Wong HT, Ardasheva A, Brumbaugh J, McLimans C, Lamendella R. Chan JP, et al. Sci Rep. 2019 Apr 3;9(1):5545. doi: 10.1038/s41598-019-41452-2. Sci Rep. 2019. PMID: 30944351 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials