Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β - PubMed (original) (raw)
. 2012 Mar 29;485(7396):123-7.
doi: 10.1038/nature11048.
Xuan Zhao, Megumi Hatori, Ruth T Yu, Grant D Barish, Michael T Lam, Ling-Wa Chong, Luciano DiTacchio, Annette R Atkins, Christopher K Glass, Christopher Liddle, Johan Auwerx, Michael Downes, Satchidananda Panda, Ronald M Evans
Affiliations
- PMID: 22460952
- PMCID: PMC3367514
- DOI: 10.1038/nature11048
Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β
Han Cho et al. Nature. 2012.
Abstract
The circadian clock acts at the genomic level to coordinate internal behavioural and physiological rhythms via the CLOCK-BMAL1 transcriptional heterodimer. Although the nuclear receptors REV-ERB-α and REV-ERB-β have been proposed to form an accessory feedback loop that contributes to clock function, their precise roles and importance remain unresolved. To establish their regulatory potential, we determined the genome-wide cis-acting targets (cistromes) of both REV-ERB isoforms in murine liver, which revealed shared recognition at over 50% of their total DNA binding sites and extensive overlap with the master circadian regulator BMAL1. Although REV-ERB-α has been shown to regulate Bmal1 expression directly, our cistromic analysis reveals a more profound connection between BMAL1 and the REV-ERB-α and REV-ERB-β genomic regulatory circuits than was previously suspected. Genes within the intersection of the BMAL1, REV-ERB-α and REV-ERB-β cistromes are highly enriched for both clock and metabolic functions. As predicted by the cistromic analysis, dual depletion of Rev-erb-α and Rev-erb-β function by creating double-knockout mice profoundly disrupted circadian expression of core circadian clock and lipid homeostatic gene networks. As a result, double-knockout mice show markedly altered circadian wheel-running behaviour and deregulated lipid metabolism. These data now unite REV-ERB-α and REV-ERB-β with PER, CRY and other components of the principal feedback loop that drives circadian expression and indicate a more integral mechanism for the coordination of circadian rhythm and metabolism.
Figures
Figure 1. Cistromic analyses of REV-ERBα and REV-ERBβ in liver
a, De novo HOMER motif analysis of in vivo REV-ERBα and REV-ERBβ binding. b, Venn diagram depicting the unique and common REV- ERBα and REV-ERBβ bound peaks. c, Commonly bound REV-ERBα and REV-ERBβ peaks are enriched for genes involved in lipid metabolism and associated with PPARs. d, REV-ERBα, REV-ERBβ and BMAL1binding at canonical circadian clock genes. Left axis indicates tag counts. e, BMAL1 cistrome significantly overlaps with REV-ERBα and REV-ERBβ. Examples of Clock related genes in overlap are listed and selected peaks shown in Supplementary Figure 4.
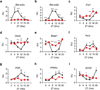
Figure 2. Circadian gene expression of many canonical core clock genes and output genes are disrupted in Livers of Rev-erbαlox/lox Rev-erbβlox/lox Albumin-Cre (L-DKO) mice
The expression levels of a, Rev-erbα, b, Rev-erbβ, c-f, canonical core clock genes (Cry1, Clock, Bmal1 and Per2) g-i, presumed output genes (PoR, PPARα and Sco2) in livers from L-DKO (Albumin-Cre positive, red labels) and wildtype (Albumin-Cre negative, black labels) mice. Livers (n=3) were harvested at each indicated ZT under 12-hour light:dark cycle. QPCR was performed in technical triplicates. Relative Unit (RU) normalized with 36B4. Error bars indicate standard error of the mean, statistical significance determined by Student t-test (* p<0.05, ** p<0.01, *** p<0.001).
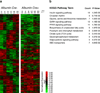
Figure 3. Broad disruption of circadian transcriptome in the absence of Rev-erbα and Rev-erbβ
a, Heatmap of genes with circadian expression in wild-type (left panel) and L-DKO (right panel) livers. 1227 unique accession numbers were selected based on fdr < 0.05. b, Genes expressed in a circadian manner that lose rhythm are highly associated with circadian and energy homeostasis functions as assessed by KEGG Pathway analysis.
Figure 4. Loss of both Rev-erbα and Rev-erbβ results in disrupted circadian wheel-running behavior and metabolic shift
a-c, Voluntary locomotor activity of wildtype, Rev-erbα−/−, Rev-erbβ−/−, and Rev-erbα−/−Rev-erbβ−/− (iDKO) mice. a, Actograms showing wheel-running activity in constant darkness after prior entrainment in light/dark. b, Activity profiles during light dark cycles. c, Chi-square periodogram of the initial 20 days in constant darkness. (n=5–9 for each mutant strain, n= 5–6 littermate controls). Representative actograms from individual mice are shown. d, Triglyceride (n=6), fasting glucose (n=6) and free fatty acid (n=6) levels in iDKO and wildtype mice. e, Respiratory exchange ratio (RER) for wildtype (black) and iDKO (red) mice (n=4). f, Model depicting the activating (Clock/BMAL1) and repressive (REV-ERBα/REV-ERBβ) transcriptional complexes whose coordinate actions generate rhythmic gene expression.
Comment in
- Drug discovery: Time in a bottle.
Bass J. Bass J. Nature. 2012 May 2;485(7396):45-6. doi: 10.1038/485045a. Nature. 2012. PMID: 22552089 No abstract available. - REV-ERBs: more than the sum of the individual parts.
Stratmann M, Schibler U. Stratmann M, et al. Cell Metab. 2012 Jun 6;15(6):791-3. doi: 10.1016/j.cmet.2012.05.006. Cell Metab. 2012. PMID: 22682217
Similar articles
- Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms.
Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Liu AC, et al. PLoS Genet. 2008 Feb 29;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. PLoS Genet. 2008. PMID: 18454201 Free PMC article. - Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists.
Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, Takahashi JS, Butler AA, Kamenecka TM, Burris TP. Solt LA, et al. Nature. 2012 Mar 29;485(7396):62-8. doi: 10.1038/nature11030. Nature. 2012. PMID: 22460951 Free PMC article. - Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour.
Banerjee S, Wang Y, Solt LA, Griffett K, Kazantzis M, Amador A, El-Gendy BM, Huitron-Resendiz S, Roberts AJ, Shin Y, Kamenecka TM, Burris TP. Banerjee S, et al. Nat Commun. 2014 Dec 23;5:5759. doi: 10.1038/ncomms6759. Nat Commun. 2014. PMID: 25536025 Free PMC article. - The REV-ERB Nuclear Receptors: Timekeepers for the Core Clock Period and Metabolism.
Adlanmerini M, Lazar MA. Adlanmerini M, et al. Endocrinology. 2023 Apr 17;164(6):bqad069. doi: 10.1210/endocr/bqad069. Endocrinology. 2023. PMID: 37149727 Free PMC article. Review. - Rev-erb alpha gives a time cue to metabolism.
Duez H, Staels B. Duez H, et al. FEBS Lett. 2008 Jan 9;582(1):19-25. doi: 10.1016/j.febslet.2007.08.032. Epub 2007 Aug 24. FEBS Lett. 2008. PMID: 17765229 Review.
Cited by
- Circadian control of hepatitis B virus replication.
Zhuang X, Forde D, Tsukuda S, D'Arienzo V, Mailly L, Harris JM, Wing PAC, Borrmann H, Schilling M, Magri A, Rubio CO, Maidstone RJ, Iqbal M, Garzon M, Minisini R, Pirisi M, Butterworth S, Balfe P, Ray DW, Watashi K, Baumert TF, McKeating JA. Zhuang X, et al. Nat Commun. 2021 Mar 12;12(1):1658. doi: 10.1038/s41467-021-21821-0. Nat Commun. 2021. PMID: 33712578 Free PMC article. - Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy.
Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MK, Paquet C, Delhaye S, Shin Y, Kamenecka TM, Schaart G, Lefebvre P, Nevière R, Burris TP, Schrauwen P, Staels B, Duez H. Woldt E, et al. Nat Med. 2013 Aug;19(8):1039-46. doi: 10.1038/nm.3213. Epub 2013 Jul 14. Nat Med. 2013. PMID: 23852339 Free PMC article. - The transcriptional repressor Rev-erbα regulates circadian expression of the astrocyte Fabp7 mRNA.
Vanderheyden WM, Fang B, Flores CC, Jager J, Gerstner JR. Vanderheyden WM, et al. Curr Res Neurobiol. 2021;2:100009. doi: 10.1016/j.crneur.2021.100009. Epub 2021 Mar 23. Curr Res Neurobiol. 2021. PMID: 34056625 Free PMC article. - Dual attenuation of proteasomal and autophagic BMAL1 degradation in Clock Δ19/+ mice contributes to improved glucose homeostasis.
Jeong K, He B, Nohara K, Park N, Shin Y, Kim S, Shimomura K, Koike N, Yoo SH, Chen Z. Jeong K, et al. Sci Rep. 2015 Jul 31;5:12801. doi: 10.1038/srep12801. Sci Rep. 2015. PMID: 26228022 Free PMC article. - Circadian topology of metabolism.
Bass J. Bass J. Nature. 2012 Nov 15;491(7424):348-56. doi: 10.1038/nature11704. Nature. 2012. PMID: 23151577 Review.
References
- Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. - PubMed
- Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. - PubMed
- Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 DK090962-02/DK/NIDDK NIH HHS/United States
- T32 HL007770/HL/NHLBI NIH HHS/United States
- R37 DK057978-34/DK/NIDDK NIH HHS/United States
- DK091618/DK/NIDDK NIH HHS/United States
- DK062434/DK/NIDDK NIH HHS/United States
- R01 HL105278-21/HL/NHLBI NIH HHS/United States
- U19 DK062434/DK/NIDDK NIH HHS/United States
- DK090962/DK/NIDDK NIH HHS/United States
- U19 DK062434-10/DK/NIDDK NIH HHS/United States
- T32-HL007770/HL/NHLBI NIH HHS/United States
- HL105278/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 HL105278/HL/NHLBI NIH HHS/United States
- T32 HL007770-15/HL/NHLBI NIH HHS/United States
- DK057978/DK/NIDDK NIH HHS/United States
- R24 DK090962/DK/NIDDK NIH HHS/United States
- R37 DK057978/DK/NIDDK NIH HHS/United States
- R01 DK091618/DK/NIDDK NIH HHS/United States
- R01 DK057978/DK/NIDDK NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials