Histone recognition and large-scale structural analysis of the human bromodomain family - PubMed (original) (raw)
. 2012 Mar 30;149(1):214-31.
doi: 10.1016/j.cell.2012.02.013.
Sarah Picaud, Maria Mangos, Tracy Keates, Jean-Philippe Lambert, Dalia Barsyte-Lovejoy, Ildiko Felletar, Rudolf Volkmer, Susanne Müller, Tony Pawson, Anne-Claude Gingras, Cheryl H Arrowsmith, Stefan Knapp
Affiliations
- PMID: 22464331
- PMCID: PMC3326523
- DOI: 10.1016/j.cell.2012.02.013
Histone recognition and large-scale structural analysis of the human bromodomain family
Panagis Filippakopoulos et al. Cell. 2012.
Abstract
Bromodomains (BRDs) are protein interaction modules that specifically recognize ε-N-lysine acetylation motifs, a key event in the reading process of epigenetic marks. The 61 BRDs in the human genome cluster into eight families based on structure/sequence similarity. Here, we present 29 high-resolution crystal structures, covering all BRD families. Comprehensive crossfamily structural analysis identifies conserved and family-specific structural features that are necessary for specific acetylation-dependent substrate recognition. Screening of more than 30 representative BRDs against systematic histone-peptide arrays identifies new BRD substrates and reveals a strong influence of flanking posttranslational modifications, such as acetylation and phosphorylation, suggesting that BRDs recognize combinations of marks rather than singly acetylated sequences. We further uncovered a structural mechanism for the simultaneous binding and recognition of diverse diacetyl-containing peptides by BRD4. These data provide a foundation for structure-based drug design of specific inhibitors for this emerging target family.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
Domain Organization, Phylogenetic Tree, and Overall Fold of BRDs (A) Domain organization of representative proteins that contain BRDs. The name and the length of the selected proteins are shown on the bar chart in the left panel. The positions of the different domains are highlighted as shown by the legend on the right. (B) Phylogenetic tree of the human BRD family. The different families are named by Roman numbers (I–VIII). Structures determined in this study, by NMR, or by other groups are indicated by blue, red, and green dots, respectively. (C) Domain flexibility as seen in the tandem BRD modules of TAF1 di-domain structure (orange PDB:
1EQF
) and a new structure (green PDB:
3UV5
), highlighting the ability of BRDs to adopt different relative orientations that may influence the recognition of their target sequences. (D) Overall structure of the BRD4(1) BRD. N and C termini and secondary structure elements are labeled. See also Figure S1.
Figure 2
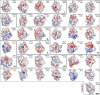
Electrostatic Surface Potential of Human BRDs The domains are grouped into the eight BRD families (shown in roman numerals). Electrostatic surface potentials are shown between −10kT/e (red) and +10kT/e (blue). The BRD names and structures (PDB accession code in black for crystal structures and red for NMR models) are shown in the figure. All domains are shown in identical orientation with their acetyl-lysine binding site facing the reader and highlighted with a dashed circle on the top-left structure (PCAF). See also Figure S3.
Figure 3
Conserved Residues and Sequence Logos Conserved residues are shown as sequence logos (lower panel in each figure), and their location is shown in the ribbon diagram above. Secondary structure elements and residue labels are colored in all figures as follows: αZ, blue; αA, pink; αB, green; αC, brown; ZA loop, magenta. Motifs and location of residues are shown for (A) ZA loop, (B) helix αA and AB loop, (C) AB loop and helix αB, and (D) helix C. The conserved Kac docking residue (N140) is shown in cyan. See also Figure S2.
Figure 4
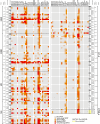
Detected Interactions of BRDs with Histones in SPOT Arrays A total of 33 BRDs were screened against an array of singly acetylated peptides that cover all possible acetylation sites in histones H1.4 (right panel), H2A, H2B, H3, and H4 (left panel). Non-Kac specific interaction (corresponding nonacetylated) peptides are shaded in gray. Spots are shaded by different spot intensities as indicated in the figure. See also Figure S4.
Figure 5
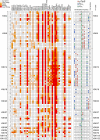
Influence of Neighboring PTMs on BRD Interaction with Histone H3 Shown are interactions detected in SPOT arrays shaded by spot intensity as indicated in the legend at the upper-left corner of the figure. The influence of lysine trimethylation (Kme3), acetylation, and phosphorylation (pT, pS) has been studied on binding to a central acetylated lysine epitope. The combination of the different modifications is indicated in the right panel. Nonmodified peptides have been included as controls to identify interactions independent of lysine acetylation. See also Figure S5.
Figure 6
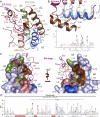
Binding of the N- and C-BRDs of BRD4 to Multiply Acetylated Histone H3 and H4 (A) Interactions detected in microSPOT arrays for BRD4(1) and BRD4(2) comprising multiply acetylated histone H3 (shown in green) or H4 (shown in blue) peptides. (B) Peptide lengths are given together with the location of the Kac marks (green hexagons). Binding of seven BRD members of the BET subfamily is summarized, highlighting the effect of multiple Kac marks as well as neighboring Ser or Thr phosphorylation. (C) Structural overlay of diacetylated peptides to the Kac binding site of BRD4(1). The binding mode is retained, although the linker between marks and the flanking residues are not the same. (D) Two representative ITC traces of polyacetylated histone H4 peptide binding to BRD4(1). Peptide sequences are shown in the inset. See also Figure S6.
Figure 7
Effect of Distance between Acetylated Lysines and In Vivo Binding of BRD4 (A) Effect of poly-Gly and linker sequence on binding of doubly acetylated peptides to BET BRDs profiled by SPOT assays. Interactions are weakened or abolished when the docking asparagine is mutated to an Ala (N140A for the first and N433A for the second BRDs of BRD4). (B) Effect of poly-Gly linker on the binding of H4K5acK8ac to BRD4(1) evaluated by ITC, demonstrating that the natural recognition sequence has the optimal sequence for binding. Peptide sequences are given in the inset. (C) The second BRD of BRD4 is more promiscuous as demonstrated by ITC, exhibiting weaker binding for all tested peptides. (D) Immunoprecipitation of Flag-tagged BRD4 from transfected cell nucleosome fraction and western blotting using anti-acetylated histone antibodies. Input represents 1% of total input. IgG was used for control immunoprecipitations. (E) Individual BRD4 BRDs purify histones with distinct acetylation status from histone fractions. Acetylated histone peptides associated with biotinylated BRD4(1) or BRD4(2) were identified by LC-MS/MS, and the relative peak intensity of individual peptides was expressed as a ratio of peak area of the specific peptide to the sum of all peak areas for acetylated histone peptides in each sample. See also Figure S7.
Figure S1
Structure-Guided Sequence Alignment of the Human Bromodomain Family, Related to Figure 1 Sequences are clustered by BRD families, highlighted by different colors corresponding to the eight families (Roman numbers I–VIII). The sequence region used for the generation of the phylogenetic tree is indicated by arrows. Location of bromodomain structural elements are shown and named on the top of the figure. Helices are indicated with solid black boxes (extracted from crystal structures) or red dashed boxes (predicted using PSIPRED) (Jones, 1999). Available representative structures (pdb-accession codes) are shown on the right (crystal structures in black and NMR models in red). The first bromodomain of BRD4 (bold and highlighted by an arrow) has been chosen as a reference sequence and the corresponding numbering is shown on top of the alignment. The red asterisk indicates a unique 17 residue insertion between helices B and C in the case of TRIM33A.
Figure S2
Structural Conservation in Helix αZ and the ZA Loop Region, Related to Figure 3 (A) Structural overview of the BRD4(1) bromodomain. Secondary structure elements and residue labels are colored as follows: αZ blue, αA pink, αB green, αC brown, ZA loop magenta. (B) Sequence conservation in helix αZ. (C) Anchoring of the ZA loop to the bromodomain core. Shown are two orientations (left and right panel) of the ZA loop and its interaction with the core structure (shown as surface). The surface is colored according to the structural elements that harbor the depicted surface residues as described in panel A. (D) Sequence conservation of for the entire family of human BRDs. Regions of low or no conservation are annotated (in pink), highlighting the advantage of employing structural and structure prediction data to define the structurally conserved BRD module.
Figure S3
Overall Structure of Human Bromodomains, Related to Figure 2 (A) Ribbon diagrams of all publicly available crystal (black labels) and NMR (red labels) structures. (B) Structural Overlay of ASH1L and the reference structure of BRD4(1). The hairpin insert, a hall mark of bromodomains of family VIII is highlighted and details are shown in the expanded view on the left panel.
Figure S4
SPOT Membrane Layout of Figure 4 and Representative Membranes for Each Family of BRDs, Related to Figure 4 Top panel: Core (H2A, H2B, H3 and H4) and linker (H1.4) histone sequences used in the SPOT membranes of Figure 4 are shown with Kac residues numbered (in the template) and highlighted (on the sequences) with a green hexagon. Boxes without a number represent control (nonacetylated) peptides for the histone marks following in the array. Lower panel: Representative membranes for each subfamily are given as follows: family I - FALZ, family II - BRDT(1), family III - EP300, family IV - BRPF1A, family V - LOC93349, family VI - MLL, family VII - TAF1L(2) and family VIII - SMARCA2A.
Figure S5
microSPOT Membrane Layout of Figure 5 and Representative Membranes for Each Family of BRDs, Related to Figure 5 Top Panel: The N-terminal sequence of human histone 3 is shown with highlighted marks that were studied for crosstalk, including lysine acetylation and (tri-)methylation as well as threonine and serine phosphorylation. Peptide sequences are given in the inset of Figure 5. The membrane layout highlights the position of central epitopes (numbered). The blow-up shows as an example the arrangement of marks around the H3K14ac epitope. Lower panel: Representative membranes for each subfamily are given as follows: family I - CECR2, family II - BRD4(2), family III - WDR9(2), family IV - BRD9, family V - BAZ2B, family VI - TRIM28, family VII - TAF1(1) and family VIII - PB1(4).
Figure S6
Binding of BRDs to Diacetylated H4 Peptides, Related to Figure 6 (A) Detail from the crystal structure of H41-11K5acK8ac binding to BRD4(1). The protein surface has been colored according to its electrostatic properties and key residues are annotated. (B) Detail of H411-21K12acK16ac binding to the surface of BRD4(1). (C) Detail of H415-25K16acK20ac binding to the surface of BRD4(1). (D) Binding of two BRD4(1) modules to the H47-17K8acK12ac peptide.
Figure S7
SPOT Membrane Layout of Figure 7 and Membranes for Each BET Bromodomain, Related to Figure 7 Membrane layout (top) of the peptides used to probe the effect on binding of a poly-glycine linker and of different residue properties between two acetyl-lysine marks on histone H4. Stained membranes of BET family bromodomains and the two inactive mutants of BRD4 are given in the lower panel.
Similar articles
- Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
Phillips M, Malone KL, Boyle BW, Montgomery C, Kressy IA, Joseph FM, Bright KM, Boyson SP, Chang S, Nix JC, Young NL, Jeffers V, Frietze S, Glass KC. Phillips M, et al. J Med Chem. 2024 May 23;67(10):8186-8200. doi: 10.1021/acs.jmedchem.4c00210. Epub 2024 May 11. J Med Chem. 2024. PMID: 38733345 Free PMC article. - The bromodomain interaction module.
Filippakopoulos P, Knapp S. Filippakopoulos P, et al. FEBS Lett. 2012 Aug 14;586(17):2692-704. doi: 10.1016/j.febslet.2012.04.045. Epub 2012 May 3. FEBS Lett. 2012. PMID: 22710155 Review. - Writers and readers of histone acetylation: structure, mechanism, and inhibition.
Marmorstein R, Zhou MM. Marmorstein R, et al. Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a018762. doi: 10.1101/cshperspect.a018762. Cold Spring Harb Perspect Biol. 2014. PMID: 24984779 Free PMC article. Review. - Cooperative binding of two acetylation marks on a histone tail by a single bromodomain.
Morinière J, Rousseaux S, Steuerwald U, Soler-López M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Müller CW, Petosa C. Morinière J, et al. Nature. 2009 Oct 1;461(7264):664-8. doi: 10.1038/nature08397. Nature. 2009. PMID: 19794495 - Emerging tools to investigate bromodomain functions.
Kougnassoukou Tchara PE, Filippakopoulos P, Lambert JP. Kougnassoukou Tchara PE, et al. Methods. 2020 Dec 1;184:40-52. doi: 10.1016/j.ymeth.2019.11.003. Epub 2019 Nov 11. Methods. 2020. PMID: 31726225 Review.
Cited by
- Current understanding of the role of the Brd4 protein in the papillomavirus lifecycle.
McBride AA, Jang MK. McBride AA, et al. Viruses. 2013 May 30;5(6):1374-94. doi: 10.3390/v5061374. Viruses. 2013. PMID: 23722886 Free PMC article. Review. - Inhibition of Bromodomain and Extraterminal Domain Family Proteins Ameliorates Experimental Renal Damage.
Suarez-Alvarez B, Morgado-Pascual JL, Rayego-Mateos S, Rodriguez RM, Rodrigues-Diez R, Cannata-Ortiz P, Sanz AB, Egido J, Tharaux PL, Ortiz A, Lopez-Larrea C, Ruiz-Ortega M. Suarez-Alvarez B, et al. J Am Soc Nephrol. 2017 Feb;28(2):504-519. doi: 10.1681/ASN.2015080910. Epub 2016 Jul 19. J Am Soc Nephrol. 2017. PMID: 27436852 Free PMC article. - Epigenetically Downregulated Breast Cancer Gene 2 through Acetyltransferase Lysine Acetyltransferase 2B Increases the Sensitivity of Colorectal Cancer to Olaparib.
Chen S, Allgayer H. Chen S, et al. Cancers (Basel). 2023 Nov 25;15(23):5580. doi: 10.3390/cancers15235580. Cancers (Basel). 2023. PMID: 38067284 Free PMC article. - NMR-based platform for fragment-based lead discovery used in screening BRD4-targeted compounds.
Yu JL, Chen TT, Zhou C, Lian FL, Tang XL, Wen Y, Shen JK, Xu YC, Xiong B, Zhang NX. Yu JL, et al. Acta Pharmacol Sin. 2016 Jul;37(7):984-93. doi: 10.1038/aps.2016.19. Epub 2016 May 30. Acta Pharmacol Sin. 2016. PMID: 27238211 Free PMC article. - Mass Spectrometry-Based Protein Footprinting Defines the Binding Pocket of Crotonylated H3K14 in the PHD1 Domain of BAF45D within the BAF Chromatin Remodeling Complex.
Martinez MR, Kiselar J, Wang B, Sadalge D, Zawadzke L, Taherbhoy A, Musser D, Davenport Y, Setser J, Chance MR, Bellon S. Martinez MR, et al. ACS Bio Med Chem Au. 2024 Jun 17;4(4):204-213. doi: 10.1021/acsbiomedchemau.4c00009. eCollection 2024 Aug 21. ACS Bio Med Chem Au. 2024. PMID: 39184054 Free PMC article.
References
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
- Beyer M., Block I., König K., Nesterov A., Fernandez S., Felgenhauer T., Schirwitz C., Leibe K., Bischoff R.F., Breitling F., Stadler V. A novel combinatorial approach to high-density peptide arrays. Methods Mol. Biol. 2009;570:309–316. - PubMed
Supplemental References
- Abagyan, R., Totrov, M., and Kuznetsov, D. (1994). ICM—a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15, 488–506.
- Akai, Y., Adachi, N., Hayashi, Y., Eitoku, M., Sano, N., Natsume, R., Kudo, N., Tanokura, M., Senda, T., and Horikoshi, M. (2010). Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction. Proc. Natl. Acad. Sci. USA 107, 8153–8158. - PMC - PubMed
- Brand, M., Rampalli, S., Chaturvedi, C.P., and Dilworth, F.J. (2008). Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat. Protoc. 3, 398–409. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases