Axonal transport deficit in a KIF5A( -/- ) mouse model - PubMed (original) (raw)
Axonal transport deficit in a KIF5A( -/- ) mouse model
Kathrin N Karle et al. Neurogenetics. 2012 May.
Abstract
Hereditary spastic paraplegia (HSP) is a neurodegenerative disorder preferentially affecting the longest corticospinal axons. More than 40 HSP genetic loci have been identified, among them SPG10, an autosomal dominant HSP caused by point mutations in the neuronal kinesin heavy chain protein KIF5A. Constitutive KIF5A knockout (KIF5A( -/- )) mice die early after birth. In these mice, lungs were unexpanded, and cell bodies of lower motor neurons in the spinal cord swollen, but the pathomechanism remained unclear. To gain insights into the pathophysiology, we characterized survival, outgrowth, and function in primary motor and sensory neuron cultures from KIF5A( -/- ) mice. Absence of KIF5A reduced survival in motor neurons, but not in sensory neurons. Outgrowth of axons and dendrites was remarkably diminished in KIF5A( -/- ) motor neurons. The number of axonal branches was reduced, whereas the number of dendrites was not altered. In KIF5A( -/- ) sensory neurons, neurite outgrowth was decreased but the number of neurites remained unchanged. In motor neurons maximum and average velocity of mitochondrial transport was reduced both in anterograde and retrograde direction. Our results point out a role of KIF5A in process outgrowth and axonal transport of mitochondria, affecting motor neurons more severely than sensory neurons. This gives pathophysiological insights into KIF5A associated HSP, and matches the clinical findings of predominant degeneration of the longest axons of the corticospinal tract.
Figures
Fig. 1
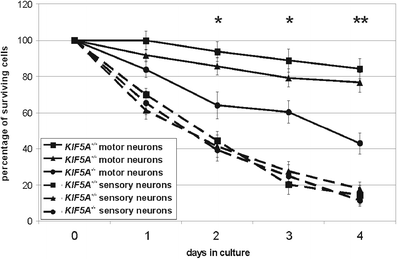
Survival of primary motor and sensory neurons from KIF5A +/+, KIF5A +/–, and KIF5A –/– mice. The number of initially counted cells was set 100 % (day 0). The percentage of surviving cells was calculated for every day in culture. Motor neurons are indicated in continuous lines, sensory neurons in broken lines. Squares show KIF5A +/+, triangles KIF5A +/–, and circles KIF5A –/– mice. After 4 days in culture mean survival of KIF5A –/– motor neurons had declined to 43 ± 6 %, whereas in KIF5A +/– 76 ± 5 %, and in KIF5A +/+ mice 84 ± 6 % of the cells survived. In sensory neurons, there was no significant difference between the three genotypes at any time point. In total, at least 11 mice were analyzed per genotype. Mean and SEM are given. * denotes significant difference with p < 0.05 between KIF5A +/+ and KIF5A –/– motor neurons, ** significant difference with p < 0.05 between KIF5A +/+ and KIF5A –/–, and KIF5A +/– and KIF5A –/– motor neurons
Fig. 2
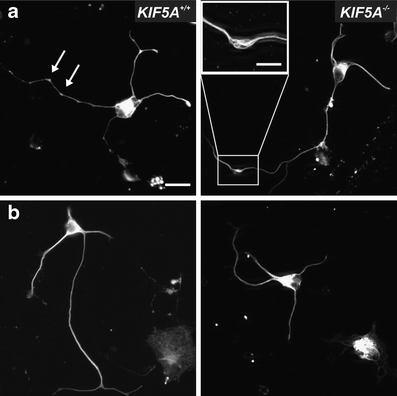
Morphology of KIF5A +/+ and KIF5A –/– motor neurons. Motor neurons were stained with an antibody against neurofilament L (white). a Shows examples with axonal swellings (arrows, magnification box), b without axonal swellings. Bar 20 μm. Bar in the magnification box 10 μm
Fig. 3
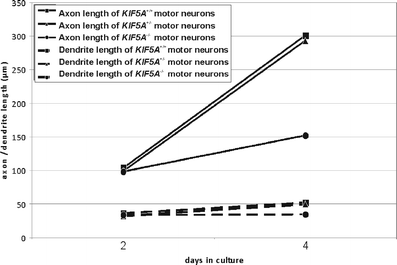
Axon and dendrite outgrowth of KIF5A +/+, KIF5A +/–, and KIF5A –/– motor neurons. Total axon length including all branches (continuous lines) and total dendrite length (broken lines) were measured after 2 and 4 days in vitro. Squares show KIF5A +/+, triangles KIF5A +/–, and circles KIF5A –/– motor neurons. Results are given as mean; due to clarity standard deviation indicators have been omitted. Measured values were linearly interpolated. For each genotype more than 150 cells from three independent experiments were analyzed
Fig. 4
Process outgrowth of motor and sensory neurons from KIF5A +/+, KIF5A +/–, and KIF5A –/– mice. For each genotype more than 150 cells from three independent experiments were analyzed. Results are given as mean and standard deviation. Total axon length including all branches (a), total dendrite length (b), number of axonal branches (c), number of dendrites (d), and cell body area (e) are given for KIF5A +/+, KIF5A +/–, and KIF5A –/– motor neurons. Neurite outgrowth (f), number of neurites (g), and cell body area (h) in KIF5A +/+, KIF5A +/–, and KIF5A –/– sensory neurons. *p < 0.05, ***p < 0.001
Fig. 5
Axonal transport of mitochondria in motor neurons. Maximum and average velocity of mitochondrial transport in anterograde (a and b) and retrograde (c and d) direction in KIF5A +/+, KIF5A +/–, and KIF5A –/– motor neurons. Mean and standard deviation are given. *p < 0.02, ***denotes p < 0.001. Results from ten independent experiments were summarized
Similar articles
- Direct evidence for axonal transport defects in a novel mouse model of mutant spastin-induced hereditary spastic paraplegia (HSP) and human HSP patients.
Kasher PR, De Vos KJ, Wharton SB, Manser C, Bennett EJ, Bingley M, Wood JD, Milner R, McDermott CJ, Miller CC, Shaw PJ, Grierson AJ. Kasher PR, et al. J Neurochem. 2009 Jul;110(1):34-44. doi: 10.1111/j.1471-4159.2009.06104.x. Epub 2009 Apr 22. J Neurochem. 2009. PMID: 19453301 - Spastic paraplegia mutation N256S in the neuronal microtubule motor KIF5A disrupts axonal transport in a Drosophila HSP model.
Füger P, Sreekumar V, Schüle R, Kern JV, Stanchev DT, Schneider CD, Karle KN, Daub KJ, Siegert VK, Flötenmeyer M, Schwarz H, Schöls L, Rasse TM. Füger P, et al. PLoS Genet. 2012;8(11):e1003066. doi: 10.1371/journal.pgen.1003066. Epub 2012 Nov 29. PLoS Genet. 2012. PMID: 23209432 Free PMC article. - KIF5A regulates axonal repair and time-dependent axonal transport of SFPQ granules and mitochondria in human motor neurons.
Guerra San Juan I, Brunner JW, Eggan K, Toonen RF, Verhage M. Guerra San Juan I, et al. Neurobiol Dis. 2025 Jan;204:106759. doi: 10.1016/j.nbd.2024.106759. Epub 2024 Dec 5. Neurobiol Dis. 2025. PMID: 39644980 - Role of kinesin-1 in the pathogenesis of SPG10, a rare form of hereditary spastic paraplegia.
Kawaguchi K. Kawaguchi K. Neuroscientist. 2013 Aug;19(4):336-44. doi: 10.1177/1073858412451655. Epub 2012 Jul 10. Neuroscientist. 2013. PMID: 22785106 Review. - Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms.
Fink JK. Fink JK. Acta Neuropathol. 2013 Sep;126(3):307-28. doi: 10.1007/s00401-013-1115-8. Epub 2013 Jul 30. Acta Neuropathol. 2013. PMID: 23897027 Free PMC article. Review.
Cited by
- Disruption of genes associated with Charcot-Marie-Tooth type 2 lead to common behavioural, cellular and molecular defects in Caenorhabditis elegans.
Soh MS, Cheng X, Vijayaraghavan T, Vernon A, Liu J, Neumann B. Soh MS, et al. PLoS One. 2020 Apr 15;15(4):e0231600. doi: 10.1371/journal.pone.0231600. eCollection 2020. PLoS One. 2020. PMID: 32294113 Free PMC article. - Impairment of Axonal Transport in Diabetes: Focus on the Putative Mechanisms Underlying Peripheral and Central Neuropathies.
Baptista FI, Pinheiro H, Gomes CA, Ambrósio AF. Baptista FI, et al. Mol Neurobiol. 2019 Mar;56(3):2202-2210. doi: 10.1007/s12035-018-1227-1. Epub 2018 Jul 12. Mol Neurobiol. 2019. PMID: 30003516 Review. - Molecular and Cellular Mechanisms Affected in ALS.
Le Gall L, Anakor E, Connolly O, Vijayakumar UG, Duddy WJ, Duguez S. Le Gall L, et al. J Pers Med. 2020 Aug 25;10(3):101. doi: 10.3390/jpm10030101. J Pers Med. 2020. PMID: 32854276 Free PMC article. Review. - Heroin Addiction Induces Axonal Transport Dysfunction in the Brain Detected by In Vivo MRI.
Luo Y, Liao C, Chen L, Zhang Y, Bao S, Deng A, Ke T, Li Q, Yang J. Luo Y, et al. Neurotox Res. 2022 Aug;40(4):1070-1085. doi: 10.1007/s12640-022-00533-3. Epub 2022 Jun 27. Neurotox Res. 2022. PMID: 35759084 - Altered CSF levels of monoamines in hereditary spastic paraparesis 10: A case series.
Andréasson M, Lagerstedt-Robinson K, Samuelsson K, Solders G, Blennow K, Paucar M, Svenningsson P. Andréasson M, et al. Neurol Genet. 2019 Jun 12;5(4):e344. doi: 10.1212/NXG.0000000000000344. eCollection 2019 Aug. Neurol Genet. 2019. PMID: 31403080 Free PMC article.
References
- Strümpell A. Beiträge zur Pathologie des Rückenmarks. I. Spastische Spinalparalysen. Archiv für Psychiatrie und Nervenkrankheiten. 1880;10:676–717. doi: 10.1007/BF02224539. - DOI
- HUGO Gene Nomenclature Committee database. http://www.genenames.org/
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases