HIF1α regulated expression of XPA contributes to cisplatin resistance in lung cancer - PubMed (original) (raw)
HIF1α regulated expression of XPA contributes to cisplatin resistance in lung cancer
Yanbin Liu et al. Carcinogenesis. 2012 Jun.
Abstract
Factors regulating nucleotide excision repair probably contribute to the heterogenous response of advanced stage lung cancer patients to drugs such as cisplatin. Studies to identify the genes in the nucleotide excision repair pathway most closely associated with resistance to cisplatin have not been conclusive. We hypothesized that Xeroderma pigmentosum complementation group A (XPA), because of its dual role in sensing and recruiting other DNA repair proteins to the damaged template, would be critical in defining sensitivity to cisplatin. Studies were conducted to identify factors regulating transcription of XPA, to assess its role in modulating sensitivity to cisplatin and its expression in primary lung tumors. Hypoxia-inducible factor 1 alpha (HIF1α) subunit was found to bind with strong affinity to a hypoxia response element sequence in the promoter of XPA. Modulating expression of HIF1α by small interfering RNA or cobalt chloride markedly reduced or increased transcription of XPA in lung cancer cell lines, respectively. Protein levels of XPA were strongly correlated with sensitivity to cisplatin (r = 0.88; P < 0.001) in cell lines and sensitivity could be increased by small interfering RNA depletion of XPA. Expression of XPA determined in 54 primary lung tumors was elevated on average 5.2-fold when compared with normal bronchial epithelial cells and correlated with levels of HIF1α (r = 0.58; P < 0.01). Together, these studies identify XPA as a novel target for regulation by HIF1α whose modulation could impact lung cancer therapy.
Figures
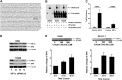
Fig. 1.
HIF1α regulated expression of XPA in lung cancer cell lines. (A) Location of HRE-binding site TACGTGC in XPA promoter relative to the transcriptional start site (CAC) and the translational start site (ATG) is shown. (B) The binding of HIF1α to HRE is demonstrated by EMSA assay. Whole cell extracts from H358 and H2228 were incubated with HRE-containing probe and then subjected to EMSA. Shift band is shown compared with the positive control (Epstein-Barr Nuclear Antigen, EBNA) in the presence of protein extract and consensus oligonucleotide for HIF1α binding (wild-type probe) compared with the scrambled probe. (C) Binding of HIF1α protein to the XPA promoter. Antibody to HIF1α was used to assess binding of this protein to the XPA promoter using EZ ChIP™. The fold enrichment of HIF1α in XPA promoter in H358 and H2228 compared with normal mouse IgG is shown. (D) Inhibition of HIF1α by small interfering RNA (siRNA) reduces expression of XPA in lung cancer cell lines. Expression levels determined by western blotting of HIF1α and XPA following HIF1α transient knockdown in H358 and H2228 are shown at indicated time points posttransfection. (E) Cobalt chloride treatment increases expression of HIF1α and XPA in lung cancer cell lines. Calu6 and SK-LU-1 cells were treated up to 48 h with 0, 100 or 500 μM cobalt chloride. Dose response increase in expression of HIF1α is shown along with increased expression of XPA seen with the 500 μM dose.
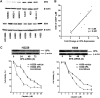
Fig. 2.
XPA protein levels in lung cancer cell lines are associated with sensitivity to cisplatin. (A) Western blot depicting protein levels of XPA in 11 lung cancer cell lines. (B) XPA expression and sensitivity to cisplatin (IC50) are highly correlated in lung cancer cell lines (r = 0.88; P < 0.001). (C) XPA knockdown sensitizes H2228 and H358 to cisplatin. Transient knockdown was confirmed by western blotting up to 72 h posttransfection. The viability of cells treated with different doses of cisplatin was measured by MTT assay. Each point represents the mean for three wells (mean ± standard error of the mean).
Fig. 3.
Expression of XPA is increased in primary lung tumors and correlates with levels of HIF1α in tumors. (A) Box plots depict the mean and standard deviation of fold increase in expression of XPA and ERCC1 compared with the average expression seen in nine NHBEC lines, HBEC1 and HBEC2 (expression level set to 1) in adenocarcinoma from smokers (Adc S) and never-smokers (Adc NS) and smokers with squamous cell carcinoma (SCC S). XPA expression is highly correlated to HIF1α in lung cancer cell lines (B) and primary tumors (C). Note that the scale for the _x_-axis differs between the three graphs.
Similar articles
- Increased levels of XPA might be the basis of cisplatin resistance in germ cell tumours.
Cierna Z, Miskovska V, Roska J, Jurkovicova D, Pulzova LB, Sestakova Z, Hurbanova L, Machalekova K, Chovanec M, Rejlekova K, Svetlovska D, Kalavska K, Kajo K, Babal P, Mardiak J, Ward TA, Mego M, Chovanec M. Cierna Z, et al. BMC Cancer. 2020 Jan 6;20(1):17. doi: 10.1186/s12885-019-6496-1. BMC Cancer. 2020. PMID: 31906898 Free PMC article. - Xeroderma Pigmentosa Group A (XPA), Nucleotide Excision Repair and Regulation by ATR in Response to Ultraviolet Irradiation.
Musich PR, Li Z, Zou Y. Musich PR, et al. Adv Exp Med Biol. 2017;996:41-54. doi: 10.1007/978-3-319-56017-5_4. Adv Exp Med Biol. 2017. PMID: 29124689 Free PMC article. Review. - XPA: DNA Repair Protein of Significant Clinical Importance.
Borszéková Pulzová L, Ward TA, Chovanec M. Borszéková Pulzová L, et al. Int J Mol Sci. 2020 Mar 22;21(6):2182. doi: 10.3390/ijms21062182. Int J Mol Sci. 2020. PMID: 32235701 Free PMC article. Review.
Cited by
- MIAT inhibits proliferation of cervical cancer cells through regulating miR-150-5p.
Liu Y, Li X, Zhang H, Huang Y. Liu Y, et al. Cancer Cell Int. 2020 Jun 15;20:242. doi: 10.1186/s12935-020-01338-0. eCollection 2020. Cancer Cell Int. 2020. PMID: 32549789 Free PMC article. - MicroRNA-326 impairs chemotherapy resistance in non small cell lung cancer by suppressing histone deacetylase SIRT1-mediated HIF1α and elevating VEGFA.
Wei J, Meng G, Wu J, Wang Y, Zhang Q, Dong T, Bao J, Wang C, Zhang J. Wei J, et al. Bioengineered. 2022 Mar;13(3):5685-5699. doi: 10.1080/21655979.2021.1993718. Bioengineered. 2022. PMID: 34696659 Free PMC article. - Impact of hypoxia on DNA repair and genome integrity.
Kaplan AR, Glazer PM. Kaplan AR, et al. Mutagenesis. 2020 Feb 13;35(1):61-68. doi: 10.1093/mutage/gez019. Mutagenesis. 2020. PMID: 31282537 Free PMC article. Review. - Oroxylin A reverses hypoxia-induced cisplatin resistance through inhibiting HIF-1α mediated XPC transcription.
Liu Y, Wang X, Li W, Xu Y, Zhuo Y, Li M, He Y, Wang X, Guo Q, Zhao L, Qiang L. Liu Y, et al. Oncogene. 2020 Nov;39(45):6893-6905. doi: 10.1038/s41388-020-01474-x. Epub 2020 Sep 25. Oncogene. 2020. PMID: 32978517 - Pharmacogenomics of cisplatin sensitivity in non-small cell lung cancer.
Rose MC, Kostyanovskaya E, Huang RS. Rose MC, et al. Genomics Proteomics Bioinformatics. 2014 Oct;12(5):198-209. doi: 10.1016/j.gpb.2014.10.003. Epub 2014 Oct 28. Genomics Proteomics Bioinformatics. 2014. PMID: 25449594 Free PMC article. Review.
References
- Helleday T, et al. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. - PubMed
- Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. - PubMed
- Morita EH, et al. Implications of the zinc-finger motif found in the DNA-binding domain of the human XPA protein. Genes Cells. 1996;1:437–442. - PubMed
- Krasikova YS, et al. Interaction of nucleotide excision repair factors XPC-HR23B, XPA, and RPA with damaged DNA. Biochemistry (Mosc.) 2008;73:886–896. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical