Balanced codon usage optimizes eukaryotic translational efficiency - PubMed (original) (raw)
Balanced codon usage optimizes eukaryotic translational efficiency
Wenfeng Qian et al. PLoS Genet. 2012.
Abstract
Cellular efficiency in protein translation is an important fitness determinant in rapidly growing organisms. It is widely believed that synonymous codons are translated with unequal speeds and that translational efficiency is maximized by the exclusive use of rapidly translated codons. Here we estimate the in vivo translational speeds of all sense codons from the budding yeast Saccharomyces cerevisiae. Surprisingly, preferentially used codons are not translated faster than unpreferred ones. We hypothesize that this phenomenon is a result of codon usage in proportion to cognate tRNA concentrations, the optimal strategy in enhancing translational efficiency under tRNA shortage. Our predicted codon-tRNA balance is indeed observed from all model eukaryotes examined, and its impact on translational efficiency is further validated experimentally. Our study reveals a previously unsuspected mechanism by which unequal codon usage increases translational efficiency, demonstrates widespread natural selection for translational efficiency, and offers new strategies to improve synthetic biology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
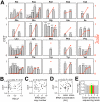
Figure 1. Relative codon selection times (_CST_s) in wild-type yeast cells in rich media.
(A) CST (grey bars) and RSCU' (orange dots) of each sense codon. _CST_s are rescaled such that the maximal observed value is 1. Error bars show one standard error, estimated by the bootstrap method. No significant negative correlation between CST and (B) RSCU', (C) tRNA gene copy number, or (D) tRNA concentration. Spearman's rank correlation coefficients (ρ) and associated P values are presented above each panel. The P value in (B) is calculated by a permutation test because of the non-independence among RSCU' values of synonymous codons. (E) No dip in RSCU' at the ribosomal A site, compared to P, E, and other neighboring sites. Geometric means of RSCU' is calculated at each codon position (as in the calculation of CAI) for ribosome profiling sequencing reads and mRNA sequencing reads, respectively; the ratio at each position is presented. Error bars show one standard error estimated by bootstrapping sequencing reads 1000 times.
Figure 2. Synonymous codons are used in proportion to cognate tRNA concentrations.
(A) Relative uses of synonymous codons in the transcriptomes of seven model eukaryotes are compared to the relative concentrations of cognate tRNAs measured from gene copy numbers, for the 12 amino acids that have at least two isoaccepting tRNA species. For comparison, genomic synonymous codon usage in S. cerevisiae is also presented. The diagonal line shows the predicted proportional relationship between tRNA concentrations and cognate codon uses that maximizes translational efficiency under tRNA shortage. (B) Euclidian and (C) Manhattan distances between the observed synonymous codon usage in S. cerevisiae and the prediction by the proportional rule are significantly smaller than chance expectations. Euclidian and Manhattan distances are defined by  and
and  , respectively, where pi and qi are codon and cognate tRNA fractions, respectively, and k is the number of different tRNA species for the amino acid concerned. The chance expectations are shown by the frequency distributions of the distances under uniformly random codon usage, determined from 106 simulations. (D) Euclidian and Manhattan distances between the observed synonymous codon usage and the predictions under the proportional rule, square rule, and truncation rule, respectively. P values indicate the probability that a distance generated by random codon usage is smaller than the observed distance, determined by 106 simulations. Log10(likelihood ratio) measures the likelihood of the proportional rule, relative to the square rule, given the actual codon usage.
, respectively, where pi and qi are codon and cognate tRNA fractions, respectively, and k is the number of different tRNA species for the amino acid concerned. The chance expectations are shown by the frequency distributions of the distances under uniformly random codon usage, determined from 106 simulations. (D) Euclidian and Manhattan distances between the observed synonymous codon usage and the predictions under the proportional rule, square rule, and truncation rule, respectively. P values indicate the probability that a distance generated by random codon usage is smaller than the observed distance, determined by 106 simulations. Log10(likelihood ratio) measures the likelihood of the proportional rule, relative to the square rule, given the actual codon usage.
Figure 3. Experimental evidence for the impact of codon usage imbalance on translational efficiency.
(A) Experimental design for examining the impact of mCherry expression on the expression of the reporter vYFP. An mCherry gene is constitutively expressed from a 2-micron plasmid in S. cerevisiae, whereas vYFP is constitutively expressed from Chromosome XII. Four different synonymous versions of mCherry are compared. (B) The codon adaptation indices (_CAI_s) of the four synonymous mCherry sequences (circled numbers), in comparison to _CAI_s of all S. cerevisiae genes. (C) Values of distance to native codon usage of yeast (D ncu) for the four mCherry sequences, in comparison to that of all S. cerevisiae genes. (D) Relationship between vYFP expression and the CAI or D ncu of mCherry, when the mCherry expression is controlled for. A finer control of mCherry expression is presented in Figure S6, where cells of the low, intermediate, and high mCherry expressions defined here are each subdivided into 5 bins. Error bars, which are barely seen, show one standard error. (E) vYFP expressions in the four strains after the removal of the plasmids that carry mCherry. Error bars show one standard error. (F) vYFP mRNA levels of the four strains relative to that of the wild-type strain, which does not carry mCherry. The mean expressions from three biological replications and the standard errors are presented.
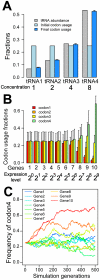
Figure 4. Computer simulation demonstrates that selection for translational efficiency results in the preferential use of codons with abundant cognate tRNAs in highly expressed genes.
Ten genes with different expression levels are considered for a haploid organism. Four synonymous codons of an amino acid are each recognized by its cognate tRNA. Concentrations of the four tRNAs differ, but the initial codon frequencies are equal. Synonymous mutations, genetic drift, and natural selection for translational efficiency are considered (see Materials and Methods). (A) Overall changes of transcriptomic codon usage averaged from 1000 simulation replications. Error bars show one standard deviation. (B) Highly expressed genes evolved stronger codon usage biases than lowly expressed genes. The averages from 1000 simulation replications are presented. Error bars show one standard deviation. (C) Evolutionary changes in the usage of codon4, the codon recognized by the most abundant tRNA, in a randomly chosen simulation replication.
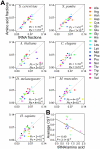
Figure 5. Amino acids are used approximately in proportion to cognate tRNA concentrations.
(A) Relative uses of amino acids estimated from the transcriptomic data of 7 model eukaryotes are compared to the relative concentrations of their cognate tRNAs measured from gene copy numbers. The diagonal line shows the predicted proportional relationship between tRNA concentrations and cognate amino acid frequencies that maximizes translational efficiency under tRNA shortage. PE (or PM) is the probability that the Euclidian (or Manhattan) distance between the amino acid usage randomly generated under a uniform distribution and that predicted by the proportional rule is smaller than the observed distance, and is estimated from 106 simulations. The distance definitions are the same as those in the legend of Figure 2, except that i is an amino acid instead of a codon. (B) The average CST of an amino acid in S. cerevisiae is negatively correlated with the availability of its cognate tRNAs, which is measured by the fraction of cognate tRNA genes among all tRNA genes divided by the frequency of the amino acid estimated from the transcriptome. The _P_-value is determined from 1000 permutations of _CST_s.
Figure 6. Similarity in transcriptomic codon usage across cell cycle stages, developmental stages, and tissues.
Distributions of pairwise Pearson's correlations of codon usage among (A) mitotic cell cycle stages in S. cerevisiae, (B) developmental stages in C. elegans, (C) tissues and developmental stages in D. melanogaster, and (D) among tissues in H. sapiens.
Figure 7. Evolutionary models of synonymous codon usage bias.
Three models that differ in the involvement of natural selection for translational accuracy and efficiency in the evolution of codon usage bias. Models I and II can be rejected by the existing data, whereas model III is supported by available data.
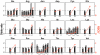
Figure 8. Matches and mismatches between preferred codons and accurate codons in S. cerevisiae.
Odds ratio (bars) measures the enrichment of a synonymous codon at evolutionarily conserved amino acid residues relative to that at non-conserved residues and is used as a proxy for translational accuracy. RSCU' (orange dots) measures the preference in codon usage. Odds ratios are estimated from either all genes (black) or the 10% most highly expressed genes (grey) of S. cerevisiae. Asterisks indicate 5% significance in the deviation of an odds ratio from 1 (uncorrected for multiple testing).
Similar articles
- Coevolution of codon usage and transfer RNA abundance.
Bulmer M. Bulmer M. Nature. 1987 Feb 19-25;325(6106):728-30. doi: 10.1038/325728a0. Nature. 1987. PMID: 2434856 - Codon usage of highly expressed genes affects proteome-wide translation efficiency.
Frumkin I, Lajoie MJ, Gregg CJ, Hornung G, Church GM, Pilpel Y. Frumkin I, et al. Proc Natl Acad Sci U S A. 2018 May 22;115(21):E4940-E4949. doi: 10.1073/pnas.1719375115. Epub 2018 May 7. Proc Natl Acad Sci U S A. 2018. PMID: 29735666 Free PMC article. - Synonymous codon ordering: a subtle but prevalent strategy of bacteria to improve translational efficiency.
Shao ZQ, Zhang YM, Feng XY, Wang B, Chen JQ. Shao ZQ, et al. PLoS One. 2012;7(3):e33547. doi: 10.1371/journal.pone.0033547. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432034 Free PMC article. - Codon usage and tRNA content in unicellular and multicellular organisms.
Ikemura T. Ikemura T. Mol Biol Evol. 1985 Jan;2(1):13-34. doi: 10.1093/oxfordjournals.molbev.a040335. Mol Biol Evol. 1985. PMID: 3916708 Review. - The Yin and Yang of codon usage.
Komar AA. Komar AA. Hum Mol Genet. 2016 Oct 1;25(R2):R77-R85. doi: 10.1093/hmg/ddw207. Epub 2016 Jun 27. Hum Mol Genet. 2016. PMID: 27354349 Free PMC article. Review.
Cited by
- Decoding properties of tRNA leave a detectable signal in codon usage bias.
Roth AC. Roth AC. Bioinformatics. 2012 Sep 15;28(18):i340-i348. doi: 10.1093/bioinformatics/bts403. Bioinformatics. 2012. PMID: 22962450 Free PMC article. - Antibody production relies on the tRNA inosine wobble modification to meet biased codon demand.
Giguère S, Wang X, Huber S, Xu L, Warner J, Weldon SR, Hu J, Phan QA, Tumang K, Prum T, Ma D, Kirsch KH, Nair U, Dedon P, Batista FD. Giguère S, et al. Science. 2024 Jan 12;383(6679):205-211. doi: 10.1126/science.adi1763. Epub 2024 Jan 11. Science. 2024. PMID: 38207021 Free PMC article. - Establishing chromosomal design-build-test-learn through a synthetic chromosome and its combinatorial reconfiguration.
Foo JL, Kitano S, Susanto AV, Jin Z, Lin Y, Luo Z, Huang L, Liang Z, Mitchell LA, Yang K, Wong A, Cai Y, Cai J, Stracquadanio G, Bader JS, Boeke JD, Dai J, Chang MW. Foo JL, et al. Cell Genom. 2023 Nov 9;3(11):100435. doi: 10.1016/j.xgen.2023.100435. eCollection 2023 Nov 8. Cell Genom. 2023. PMID: 38020970 Free PMC article. - Rate-limiting steps in yeast protein translation.
Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Shah P, et al. Cell. 2013 Jun 20;153(7):1589-601. doi: 10.1016/j.cell.2013.05.049. Cell. 2013. PMID: 23791185 Free PMC article. - Full-length ribosome density prediction by a multi-input and multi-output model.
Tian T, Li S, Lang P, Zhao D, Zeng J. Tian T, et al. PLoS Comput Biol. 2021 Mar 26;17(3):e1008842. doi: 10.1371/journal.pcbi.1008842. eCollection 2021 Mar. PLoS Comput Biol. 2021. PMID: 33770074 Free PMC article.
References
- Hershberg R, Petrov DA. General rules for optimal codon choice. PLoS Genet. 2009;5:e1000556. doi: 10.1371/journal.pgen.1000556. - DOI - PMC - PubMed
- Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, et al. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16:8207–8211. - PMC - PubMed
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. - PubMed
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases