Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA - PubMed (original) (raw)
Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA
Susanna Sawyer et al. PLoS One. 2012.
Abstract
DNA that survives in museum specimens, bones and other tissues recovered by archaeologists is invariably fragmented and chemically modified. The extent to which such modifications accumulate over time is largely unknown but could potentially be used to differentiate between endogenous old DNA and present-day DNA contaminating specimens and experiments. Here we examine mitochondrial DNA sequences from tissue remains that vary in age between 18 and 60,000 years with respect to three molecular features: fragment length, base composition at strand breaks, and apparent C to T substitutions. We find that fragment length does not decrease consistently over time and that strand breaks occur preferentially before purine residues by what may be at least two different molecular mechanisms that are not yet understood. In contrast, the frequency of apparent C to T substitutions towards the 5'-ends of molecules tends to increase over time. These nucleotide misincorporations are thus a useful tool to distinguish recent from ancient DNA sources in specimens that have not been subjected to unusual or harsh treatments.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
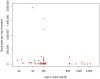
Figure 1. Amounts of endogenous mtDNA sequences (bp) determined per milligram (mg) of tissue as a function of age.
Note that since the Neandertal specimens were all ascertained for containing endogenous DNA they are excluded from this analysis. Nine samples known to have been “roasted” over fire and treated with ponal glue are indicated by crosses and four samples treated by the “Leipzig cocktail” are indicated by circles.
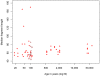
Figure 2. Median length of endogenous mtDNA fragment as a function of age.
Nine samples known to have been “roasted” over fire and treated with ponal glue are indicated by crosses and four samples treated by the “Leipzig cocktail” are indicated by circles.
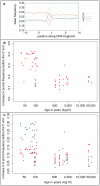
Figure 3. Characteristics of purine frequency prior to strand breaks.
A: Base frequencies 5′ and 3′ of 5′-ends of endogenous mtDNA fragments of a horse (sample 54). B: Increase in purine frequency at position immediately 5′ (position −1) of mtDNA fragment ends relative to positions −5 to −10 as a function of age. Nine samples known to have been “roasted” over fire and treated with ponal glue are indicated by crosses and four samples treated by the “Leipzig cocktail” are indicated by circles. C: Increase in A (blue) and G (red) frequencies at position −1 of mtDNA fragment. Nine samples known to have been “roasted” over fire and treated with ponal glue are indicated by crosses and four samples treated by the “Leipzig cocktail” are indicated by circles.
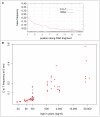
Figure 4. Characteristics of C to T misincorporations.
A: C to T misincorporations at the first 15 bases of endogenous mtDNA fragments from of a 500–600 year old horse sample (sample 54). B: C to T misincorporations at the first position of mtDNA fragments as a function of age. Nine samples known to have been “roasted” over fire and treated with ponal glue are indicated by crosses and four samples treated by the “Leipzig cocktail” are indicated by circles.
Similar articles
- Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA.
Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, Ovodov ND, Antipina EE, Baryshnikov GF, Kuzmin YV, Vasilevski AA, Wuenschell GE, Termini J, Hofreiter M, Jaenicke-Després V, Pääbo S. Stiller M, et al. Proc Natl Acad Sci U S A. 2006 Sep 12;103(37):13578-84. doi: 10.1073/pnas.0605327103. Epub 2006 Aug 25. Proc Natl Acad Sci U S A. 2006. PMID: 16938852 Free PMC article. - Patterns of damage in genomic DNA sequences from a Neandertal.
Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, Lachmann M, Pääbo S. Briggs AW, et al. Proc Natl Acad Sci U S A. 2007 Sep 11;104(37):14616-21. doi: 10.1073/pnas.0704665104. Epub 2007 Aug 21. Proc Natl Acad Sci U S A. 2007. PMID: 17715061 Free PMC article. - A complete mtDNA genome of an early modern human from Kostenki, Russia.
Krause J, Briggs AW, Kircher M, Maricic T, Zwyns N, Derevianko A, Pääbo S. Krause J, et al. Curr Biol. 2010 Feb 9;20(3):231-6. doi: 10.1016/j.cub.2009.11.068. Epub 2009 Dec 31. Curr Biol. 2010. PMID: 20045327 - Genetic analyses from ancient DNA.
Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Pääbo S, et al. Annu Rev Genet. 2004;38:645-79. doi: 10.1146/annurev.genet.37.110801.143214. Annu Rev Genet. 2004. PMID: 15568989 Review. - Ancient DNA studies: new perspectives on old samples.
Rizzi E, Lari M, Gigli E, De Bellis G, Caramelli D. Rizzi E, et al. Genet Sel Evol. 2012 Jul 6;44(1):21. doi: 10.1186/1297-9686-44-21. Genet Sel Evol. 2012. PMID: 22697611 Free PMC article. Review.
Cited by
- An easy-to-use pipeline to analyze amplicon-based Next Generation Sequencing results of human mitochondrial DNA from degraded samples.
Cuesta-Aguirre DR, Malgosa A, Santos C. Cuesta-Aguirre DR, et al. PLoS One. 2024 Nov 21;19(11):e0311115. doi: 10.1371/journal.pone.0311115. eCollection 2024. PLoS One. 2024. PMID: 39570888 Free PMC article. - SAFARI: Pangenome Alignment of Ancient DNA Using Purine/Pyrimidine Encodings.
Rubin J, van Waaij J, Kraft L, Sirén J, Sackett PW, Renaud G. Rubin J, et al. bioRxiv [Preprint]. 2024 Oct 8:2024.08.12.607489. doi: 10.1101/2024.08.12.607489. bioRxiv. 2024. PMID: 39415996 Free PMC article. Preprint. - Improved detection of methylation in ancient DNA.
Sawyer S, Gelabert P, Yakir B, Llanos-Lizcano A, Sperduti A, Bondioli L, Cheronet O, Neugebauer-Maresch C, Teschler-Nicola M, Novak M, Pap I, Szikossy I, Hajdu T, Moiseyev V, Gromov A, Zariņa G, Meshorer E, Carmel L, Pinhasi R. Sawyer S, et al. Genome Biol. 2024 Oct 10;25(1):261. doi: 10.1186/s13059-024-03405-5. Genome Biol. 2024. PMID: 39390557 Free PMC article. - Life history and ancestry of the late Upper Palaeolithic infant from Grotta delle Mura, Italy.
Higgins OA, Modi A, Cannariato C, Diroma MA, Lugli F, Ricci S, Zaro V, Vai S, Vazzana A, Romandini M, Yu H, Boschin F, Magnone L, Rossini M, Di Domenico G, Baruffaldi F, Oxilia G, Bortolini E, Dellù E, Moroni A, Ronchitelli A, Talamo S, Müller W, Calattini M, Nava A, Posth C, Lari M, Bondioli L, Benazzi S, Caramelli D. Higgins OA, et al. Nat Commun. 2024 Sep 20;15(1):8248. doi: 10.1038/s41467-024-51150-x. Nat Commun. 2024. PMID: 39304646 Free PMC article. - Maximizing efficiency in sedimentary ancient DNA analysis: a novel extract pooling approach.
Oberreiter V, Gelabert P, Brück F, Franz S, Zelger E, Szedlacsek S, Cheronet O, Cano FT, Exler F, Zagorc B, Karavanić I, Banda M, Gasparyan B, Straus LG, Gonzalez Morales MR, Kappelman J, Stahlschmidt M, Rattei T, Kraemer SM, Sawyer S, Pinhasi R. Oberreiter V, et al. Sci Rep. 2024 Aug 20;14(1):19388. doi: 10.1038/s41598-024-69741-5. Sci Rep. 2024. PMID: 39169089 Free PMC article.
References
- Paabo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, et al. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–679. - PubMed
- Noonan JP, Hofreiter M, Smith D, Priest JR, Rohland N, et al. Genomic sequencing of Pleistocene cave bears. Science. 2005;309:597–599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources