GPS-ARM: computational analysis of the APC/C recognition motif by predicting D-boxes and KEN-boxes - PubMed (original) (raw)
GPS-ARM: computational analysis of the APC/C recognition motif by predicting D-boxes and KEN-boxes
Zexian Liu et al. PLoS One. 2012.
Abstract
Anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase incorporated with Cdh1 and/or Cdc20 recognizes and interacts with specific substrates, and faithfully orchestrates the proper cell cycle events by targeting proteins for proteasomal degradation. Experimental identification of APC/C substrates is largely dependent on the discovery of APC/C recognition motifs, e.g., the D-box and KEN-box. Although a number of either stringent or loosely defined motifs proposed, these motif patterns are only of limited use due to their insufficient powers of prediction. We report the development of a novel GPS-ARM software package which is useful for the prediction of D-boxes and KEN-boxes in proteins. Using experimentally identified D-boxes and KEN-boxes as the training data sets, a previously developed GPS (Group-based Prediction System) algorithm was adopted. By extensive evaluation and comparison, the GPS-ARM performance was found to be much better than the one using simple motifs. With this powerful tool, we predicted 4,841 potential D-boxes in 3,832 proteins and 1,632 potential KEN-boxes in 1,403 proteins from H. sapiens, while further statistical analysis suggested that both the D-box and KEN-box proteins are involved in a broad spectrum of biological processes beyond the cell cycle. In addition, with the co-localization information, we predicted hundreds of mitosis-specific APC/C substrates with high confidence. As the first computational tool for the prediction of APC/C-mediated degradation, GPS-ARM is a useful tool for information to be used in further experimental investigations. The GPS-ARM is freely accessible for academic researchers at: http://arm.biocuckoo.org.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
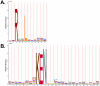
Figure 1. The Sequence logos of ARM(2, 6) and ARM(8, 15) were generated by the HMM-Logo (LogoMat-M) for the (A) D-box and (B) KEN-box, respectively.
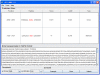
Figure 2. Screen snapshot of the GPS-ARM 1.0 software.
The default thresholds were chosen for the D-box (high) and KEN-box (low). As an example, the prediction results for the human centromere protein F/CENP-F (UniProt ID: P49454) are shown.
Figure 3. The prediction performance of GPS-ARM 1.0.
The LOO validation and 4-, 6-, 8- and 10-fold cross-validations were performed for (A) the D-box and (B) the KEN-box.
Similar articles
- Structural analysis of human Cdc20 supports multisite degron recognition by APC/C.
Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Tian W, et al. Proc Natl Acad Sci U S A. 2012 Nov 6;109(45):18419-24. doi: 10.1073/pnas.1213438109. Epub 2012 Oct 22. Proc Natl Acad Sci U S A. 2012. PMID: 23091007 Free PMC article. - The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C).
Sczaniecka M, Feoktistova A, May KM, Chen JS, Blyth J, Gould KL, Hardwick KG. Sczaniecka M, et al. J Biol Chem. 2008 Aug 22;283(34):23039-47. doi: 10.1074/jbc.M803594200. Epub 2008 Jun 13. J Biol Chem. 2008. PMID: 18556659 Free PMC article. - Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes.
Zur A, Brandeis M. Zur A, et al. EMBO J. 2002 Sep 2;21(17):4500-10. doi: 10.1093/emboj/cdf452. EMBO J. 2002. PMID: 12198152 Free PMC article. - Structural insights into anaphase-promoting complex function and mechanism.
Barford D. Barford D. Philos Trans R Soc Lond B Biol Sci. 2011 Dec 27;366(1584):3605-24. doi: 10.1098/rstb.2011.0069. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 22084387 Free PMC article. Review. - Structure, function and mechanism of the anaphase promoting complex (APC/C).
Barford D. Barford D. Q Rev Biophys. 2011 May;44(2):153-90. doi: 10.1017/S0033583510000259. Epub 2010 Nov 22. Q Rev Biophys. 2011. PMID: 21092369 Review.
Cited by
- Kaposi's Sarcoma-Associated Herpesvirus Lytic Replication Is Independent of Anaphase-Promoting Complex Activity.
Elbasani E, Gramolelli S, Günther T, Gabaev I, Grundhoff A, Ojala PM. Elbasani E, et al. J Virol. 2020 Jun 16;94(13):e02079-19. doi: 10.1128/JVI.02079-19. Print 2020 Jun 16. J Virol. 2020. PMID: 32295923 Free PMC article. - MiCroKiTS 4.0: a database of midbody, centrosome, kinetochore, telomere and spindle.
Huang Z, Ma L, Wang Y, Pan Z, Ren J, Liu Z, Xue Y. Huang Z, et al. Nucleic Acids Res. 2015 Jan;43(Database issue):D328-34. doi: 10.1093/nar/gku1125. Epub 2014 Nov 11. Nucleic Acids Res. 2015. PMID: 25392421 Free PMC article. - Counting Degrons: Lessons From Multivalent Substrates for Targeted Protein Degradation.
Okoye CN, Rowling PJE, Itzhaki LS, Lindon C. Okoye CN, et al. Front Physiol. 2022 Jul 4;13:913063. doi: 10.3389/fphys.2022.913063. eCollection 2022. Front Physiol. 2022. PMID: 35860655 Free PMC article. Review. - Differential Expression of Genes for Ubiquitin Ligases in Medulloblastoma Subtypes.
Vriend J, Tate RB. Vriend J, et al. Cerebellum. 2019 Jun;18(3):469-488. doi: 10.1007/s12311-019-1009-y. Cerebellum. 2019. PMID: 30810905 - Degradation of Ndd1 by APC/C(Cdh1) generates a feed forward loop that times mitotic protein accumulation.
Sajman J, Zenvirth D, Nitzan M, Margalit H, Simpson-Lavy KJ, Reiss Y, Cohen I, Ravid T, Brandeis M. Sajman J, et al. Nat Commun. 2015 May 11;6:7075. doi: 10.1038/ncomms8075. Nat Commun. 2015. PMID: 25959309
References
- Milestones in cell division. Nat Cell Biol. 2001;3:E265. - PubMed
- Nasmyth K. A prize for proliferation. Cell. 2001;107:689–701. - PubMed
- Barford D. Structure, function and mechanism of the anaphase promoting complex (APC/C). Q Rev Biophys. 2011;44:153–190. - PubMed
- Thornton BR, Toczyski DP. Precise destruction: an emerging picture of the APC. Genes Dev. 2006;20:3069–3078. - PubMed
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous