IFITM proteins restrict antibody-dependent enhancement of dengue virus infection - PubMed (original) (raw)
IFITM proteins restrict antibody-dependent enhancement of dengue virus infection
Ying Kai Chan et al. PLoS One. 2012.
Abstract
Interferon-inducible transmembrane (IFITM) proteins restrict the entry processes of several pathogenic viruses, including the flaviviruses West Nile virus and dengue virus (DENV). DENV infects cells directly or via antibody-dependent enhancement (ADE) in Fc-receptor-bearing cells, a process thought to contribute to severe disease in a secondary infection. Here we investigated whether ADE-mediated DENV infection bypasses IFITM-mediated restriction or whether IFITM proteins can be protective in a secondary infection. We observed that IFITM proteins restricted ADE-mediated and direct infection with comparable efficiencies in a myelogenous leukemia cell line. Our data suggest that IFITM proteins can contribute to control of secondary DENV infections.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
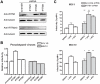
Figure 1. IFITM proteins restrict both direct and ADE-mediated DENV infection of K562 cells.
(A) K562 cells were treated with the indicated concentrations of human interferon-α or mock treated, and lysed for Western blot analysis 48 h later, using an antibody that recognizes IFITM1 alone, or one that recognizes both IFITM2 and IFITM3. (B) K562 cells were stably transduced to express human IFITM1, 2, or 3 or with the control vector pQCXIP. IFITM protein expression was measured by Western blot using an anti-c-myc antibody. (C) Stably transduced K562 cells characterized in panel B were infected with an MLV-based retrovirus expressing EGFP and pseudotyped with MACV or IAV H5 entry proteins, as previously described . Infection was determined by flow cytometry, and normalized to K562 cells transduced with vector alone. (D) K562 cells characterized in panel B were infected with infectious DENV2 NGC strain (labeled “DENV2” for virus only) at the indicated MOI, or the same amount of infectious DENV2 pre-opsonized with enhancing titers of antibodies against DENV structural proteins prM or E (“+2H2” and “+4G2” respectively) , . Cells were washed after 1.5 h and incubated for ∼24 h. Intracellular staining of DENV antigen was performed with a DyLight-649-conjugated antibody against prM and infection was determined by flow cytometry. Experiment is representative of three with similar results. (E) An experiment similar to that shown in panel D was performed except that infection was assayed by measuring viral loads in the supernatant by plaque assays using BHK cells. (F) An experiment similar to that in panel D was performed, except that J774A.1 murine macrophage cells were stably transduced to express murine orthologs of IFITM1, 2, or 3 or with the control vector pQCXIP. Stably transduced cells were infected with infectious DENV2 NGC strain at ∼MOI 5 and incubated for ∼2 days before harvesting for flow cytometry. Error bars indicate standard error. Single and double asterisks indicate statistically significant (P<0.05 and P<0.005, respectively) differences between IFITM protein expressing and control cells for corresponding infection conditions.
Figure 2. Endogenous IFITM1 restricts both direct and ADE-mediated DENV infection of K562 cells.
(A) K562 cells were stably transduced to express shRNA targeting IFITM1, 2, or 3 or non-targeting control shRNA (scrambled). IFITM protein expression was measured by Western blot, as in Fig. 1A. (B) shRNA-transduced K562 cells characterized in panel A were infected with the indicated pseudoviruses as described in Fig. 1C. (C) shRNA-transduced K562 cells characterized in panel A were infected with infectious DENV2 NGC strain as described in Fig. 1D. Error bars indicate standard error. Single and double asterisks indicate statistically significant (P<0.05 and P<0.005, respectively) differences between cells expressing IFITM1 and control shRNA for corresponding infection conditions. Experiment is representative of three with similar results.
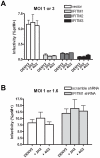
Figure 3. IFITM proteins restrict direct and ADE-mediated infection with similar efficiencies.
(A) K562 cells stably transduced to express human IFITM1, 2, or 3 or with control vector were infected as in Fig. 1D, except that an MOI of 3.0 was used for non-opsonized DENV, and an MOI of 1.0 was used for DENV pre-opsonized with the antibodies 2H2 or 4G2. Different MOIs were used to achieve comparable infection levels between direct and ADE-mediated infection. (B) K562 cells stably transduced to express control shRNA or shRNA targeting IFITM1 were infected as described in Fig. 2C, except that an MOI of 1.6 was used for non-opsonized DENV, and an MOI of 1.0 was used for DENV pre-opsonized with the indicated antibodies. Standard error bars are shown. For each transduction condition, differences between direct infection and ADE-mediated infection were not statistically significant.
Similar articles
- Modulation of Dengue/Zika Virus Pathogenicity by Antibody-Dependent Enhancement and Strategies to Protect Against Enhancement in Zika Virus Infection.
Khandia R, Munjal A, Dhama K, Karthik K, Tiwari R, Malik YS, Singh RK, Chaicumpa W. Khandia R, et al. Front Immunol. 2018 Apr 23;9:597. doi: 10.3389/fimmu.2018.00597. eCollection 2018. Front Immunol. 2018. PMID: 29740424 Free PMC article. Review. - The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus.
Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. Brass AL, et al. Cell. 2009 Dec 24;139(7):1243-54. doi: 10.1016/j.cell.2009.12.017. Cell. 2009. PMID: 20064371 Free PMC article. - Antibody with an engineered Fc region as a therapeutic agent against dengue virus infection.
Ramadhany R, Hirai I, Sasaki T, Ono K, Ramasoota P, Ikuta K, Kurosu T. Ramadhany R, et al. Antiviral Res. 2015 Dec;124:61-8. doi: 10.1016/j.antiviral.2015.10.012. Epub 2015 Oct 30. Antiviral Res. 2015. PMID: 26522769 - Dengue virus compartmentalization during antibody-enhanced infection.
Ong EZ, Zhang SL, Tan HC, Gan ES, Chan KR, Ooi EE. Ong EZ, et al. Sci Rep. 2017 Jan 13;7:40923. doi: 10.1038/srep40923. Sci Rep. 2017. PMID: 28084461 Free PMC article. - Paradoxical role of antibodies in dengue virus infections: considerations for prophylactic vaccine development.
Acosta EG, Bartenschlager R. Acosta EG, et al. Expert Rev Vaccines. 2016;15(4):467-82. doi: 10.1586/14760584.2016.1121814. Epub 2015 Dec 15. Expert Rev Vaccines. 2016. PMID: 26577689 Review.
Cited by
- Cross Talk between MicroRNAs and Dengue Virus.
Macha NO, Komarasamy TV, Harun S, Adnan NAA, Hassan SS, Balasubramaniam VRMT. Macha NO, et al. Am J Trop Med Hyg. 2024 Apr 2;110(5):856-867. doi: 10.4269/ajtmh.23-0546. Print 2024 May 1. Am J Trop Med Hyg. 2024. PMID: 38579704 Review. - Role of interferon-induced transmembrane protein family in cancer progression: a special focus on pancreatic cancer.
Wang P, Pan Y, Zhang Y, Chen C, Hu J, Wang X. Wang P, et al. Med Oncol. 2024 Mar 12;41(4):85. doi: 10.1007/s12032-024-02308-6. Med Oncol. 2024. PMID: 38472606 Review. - Role of Viral Envelope Proteins in Determining Susceptibility of Viruses to IFITM Proteins.
Marceau T, Braibant M. Marceau T, et al. Viruses. 2024 Feb 5;16(2):254. doi: 10.3390/v16020254. Viruses. 2024. PMID: 38400030 Free PMC article. Review. - Ethyl Gallate Inhibits Bovine Viral Diarrhea Virus by Promoting IFITM3 Expression, Lysosomal Acidification and Protease Activity.
Zhang L, Yang G, Wang J, Zhang J, Chen K, Xiong X, Zhu Y, Xu C, Wang J. Zhang L, et al. Int J Mol Sci. 2023 May 12;24(10):8637. doi: 10.3390/ijms24108637. Int J Mol Sci. 2023. PMID: 37239983 Free PMC article. - IFITM2 Presents Antiviral Response through Enhancing Type I IFN Signaling Pathway.
Chen L, Li X, Deng Y, Bi Y, Yan Z, Yang Y, Zhang X, Li H, Xie J, Feng R. Chen L, et al. Viruses. 2023 Mar 28;15(4):866. doi: 10.3390/v15040866. Viruses. 2023. PMID: 37112847 Free PMC article.
References
- Halstead SB. Dengue. Lancet. 2007;370:1644–1652. - PubMed
- Jessie K, Fong MY, Devi S, Lam SK, Wong KT. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J Infect Dis. 2004;189:1411–1418. - PubMed
- Kliks SC, Nimmanitya S, Nisalak A, Burke DS. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am J Trop Med Hyg. 1988;38:411–419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RR000168/RR/NCRR NIH HHS/United States
- U54 AI057159/AI/NIAID NIH HHS/United States
- K99 AI093679/AI/NIAID NIH HHS/United States
- R00 AI093679/AI/NIAID NIH HHS/United States
- P51 RR000168/RR/NCRR NIH HHS/United States
- K26 RR000168/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases