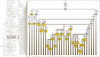A "Copernican" reassessment of the human mitochondrial DNA tree from its root - PubMed (original) (raw)
A "Copernican" reassessment of the human mitochondrial DNA tree from its root
Doron M Behar et al. Am J Hum Genet. 2012.
Erratum in
- Am J Hum Genet. 2012 May 4;90(5):936
Abstract
Mutational events along the human mtDNA phylogeny are traditionally identified relative to the revised Cambridge Reference Sequence, a contemporary European sequence published in 1981. This historical choice is a continuous source of inconsistencies, misinterpretations, and errors in medical, forensic, and population genetic studies. Here, after having refined the human mtDNA phylogeny to an unprecedented level by adding information from 8,216 modern mitogenomes, we propose switching the reference to a Reconstructed Sapiens Reference Sequence, which was identified by considering all available mitogenomes from Homo neanderthalensis. This "Copernican" reassessment of the human mtDNA tree from its deepest root should resolve previous problems and will have a substantial practical and educational influence on the scientific and public perception of human evolution by clarifying the core principles of common ancestry for extant descendants.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
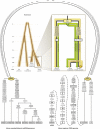
Figure 1
Schematic Representation of the Human mtDNA Phylogeny within Hominini (Left) Hominini phylogeny illustrating approximate divergence times of the studied species. The positions of the RSRS and the putative Reconstructed Neanderthal Reference Sequence (RNRS) are shown. (Right) Magnification of the human mtDNA phylogeny. Mutated nucleotide positions separating the nodes of the two basal human haplogroups L0 and L1′2′3′4′5′6 and their derived states as compared to the RSRS are shown. The positions of the rCRS and the RSRS are indicated by golden and a green five-pointed stars, respectively. Accordingly, the number of mutations counted from the rCRS (NC_012920) or the RSRS (Sequence S1) to the L0d1c1b (EU092832) and H4a1a (HQ860291) haplotypes retrieved from a San and a German, respectively, are marked on the golden and green branches. The principle of equidistant star-like radiation from the common ancestor of all contemporary haplotypes is highlighted when the RSRS is preferred over the rCRS as the reference sequence.
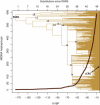
Figure 2
Human mtDNA Phylogeny A schematic representation of the most parsimonious human mtDNA phylogeny inferred from 18,843 complete mtDNA sequences with the structure shown explicitly for bifurcations that occurred 40,000 years before present (YBP) or earlier, and a graph showing the explosion of haplogroups since then. The y axis indicates the approximate number of haplogroups from each time layer that have survived to nowadays. The upper and lower x axes of the rooted tree are scaled according to the number of accumulated mutations since the RSRS and the corresponding coalescence ages, respectively.
Figure 3
Haplogroup H4 internal cladistic structure (Left) Haplogroup H4 as first reported. Mutations in bold were considered diagnostic for the haplogroup. (Right) Haplogroup H4 as currently resolved with a total of 236 H4 mitogenomes. An almost perfect resolution of the nested hierarchy is achieved. Additional haplogroups suggested herein are shown in yellow. Control-region mutations are noted in blue.
Similar articles
- Phylogenomic study and classification of mitochondrial DNA through virtual genomic fingerprints.
Jaimes Díaz H, Martínez Covarrubias EI, Murcia Garzón JE, Flores Valdez M, Muñoz Ramírez ZY, Ramírez Calzada CA, Bohra R, Méndez Tenorio A. Jaimes Díaz H, et al. Mitochondrion. 2021 Mar;57:294-299. doi: 10.1016/j.mito.2020.11.016. Epub 2020 Dec 8. Mitochondrion. 2021. PMID: 33301927 - Updating the African human mitochondrial DNA tree: Relevance to forensic and population genetics.
Heinz T, Pala M, Gómez-Carballa A, Richards MB, Salas A. Heinz T, et al. Forensic Sci Int Genet. 2017 Mar;27:156-159. doi: 10.1016/j.fsigen.2016.12.016. Epub 2016 Dec 31. Forensic Sci Int Genet. 2017. PMID: 28086175 - The case for the continuing use of the revised Cambridge Reference Sequence (rCRS) and the standardization of notation in human mitochondrial DNA studies.
Bandelt HJ, Kloss-Brandstätter A, Richards MB, Yao YG, Logan I. Bandelt HJ, et al. J Hum Genet. 2014 Feb;59(2):66-77. doi: 10.1038/jhg.2013.120. Epub 2013 Dec 5. J Hum Genet. 2014. PMID: 24304692 Review. - Archaic human genomics.
Disotell TR. Disotell TR. Am J Phys Anthropol. 2012;149 Suppl 55:24-39. doi: 10.1002/ajpa.22159. Epub 2012 Nov 2. Am J Phys Anthropol. 2012. PMID: 23124308 Review.
Cited by
- Pervasive findings of directional selection realize the promise of ancient DNA to elucidate human adaptation.
Akbari A, Barton AR, Gazal S, Li Z, Kariminejad M, Perry A, Zeng Y, Mittnik A, Patterson N, Mah M, Zhou X, Price AL, Lander ES, Pinhasi R, Rohland N, Mallick S, Reich D. Akbari A, et al. bioRxiv [Preprint]. 2024 Sep 15:2024.09.14.613021. doi: 10.1101/2024.09.14.613021. bioRxiv. 2024. PMID: 39314480 Free PMC article. Preprint. - Association analysis of mitochondrial DNA heteroplasmic variants: Methods and application.
Sun X, Bulekova K, Yang J, Lai M, Pitsillides AN, Liu X, Zhang Y, Guo X, Yong Q, Raffield LM, Rotter JI, Rich SS, Abecasis G, Carson AP, Vasan RS, Bis JC, Psaty BM, Boerwinkle E, Fitzpatrick AL, Satizabal CL, Arking DE, Ding J, Levy D; TOPMed mtDNA working group; Liu C. Sun X, et al. Mitochondrion. 2024 Nov;79:101954. doi: 10.1016/j.mito.2024.101954. Epub 2024 Sep 7. Mitochondrion. 2024. PMID: 39245194 - Mitochondrial DNA Genomes Reveal Relaxed Purifying Selection During Human Population Expansion after the Last Glacial Maximum.
Zheng HX, Yan S, Zhang M, Gu Z, Wang J, Jin L. Zheng HX, et al. Mol Biol Evol. 2024 Sep 4;41(9):msae175. doi: 10.1093/molbev/msae175. Mol Biol Evol. 2024. PMID: 39162340 Free PMC article. - mtDNA "nomenclutter" and its consequences on the interpretation of genetic data.
Bajić V, Schulmann VH, Nowick K. Bajić V, et al. BMC Ecol Evol. 2024 Aug 19;24(1):110. doi: 10.1186/s12862-024-02288-1. BMC Ecol Evol. 2024. PMID: 39160470 Free PMC article. - Long shared haplotypes identify the Southern Urals as a primary source for the 10th century Hungarians.
Gyuris B, Vyazov L, Türk A, Flegontov P, Szeifert B, Langó P, Mende BG, Csáky V, Chizhevskiy AA, Gazimzyanov IR, Khokhlov AA, Kolonskikh AG, Matveeva NP, Ruslanova RR, Rykun MP, Sitdikov A, Volkova EV, Botalov SG, Bugrov DG, Grudochko IV, Komar O, Krasnoperov AA, Poshekhonova OE, Chikunova I, Sungatov F, Stashenkov DA, Zubov S, Zelenkov AS, Ringbauer H, Cheronet O, Pinhasi R, Akbari A, Rohland N, Mallick S, Reich D, Szécsényi-Nagy A. Gyuris B, et al. bioRxiv [Preprint]. 2024 Jul 23:2024.07.21.599526. doi: 10.1101/2024.07.21.599526. bioRxiv. 2024. PMID: 39091721 Free PMC article. Preprint.
References
- Darwin C. John Murray; London: 1859. Natural Selection. On the Origin of Species by Means of Natural Selection, or, The Preservation of Favoured Races in the Struggle for Life, Chapter 4. - PubMed
- Delsuc F., Brinkmann H., Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005;6:361–375. - PubMed
- Kivisild T., Metspalu E., Bandelt H.J., Richards M., Villems R. The world mtDNA phylogeny. In: Bandelt H.J., Macaulay V., Richards M., editors. Human mitochondrial DNA and the evolution of Homo sapiens. Springer-Verlag; Berlin: 2006. pp. 149–179.
- Ingman M., Kaessmann H., Pääbo S., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources