The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription - PubMed (original) (raw)
The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription
Min Liu et al. Mol Cell. 2012.
Abstract
Super elongation complexes (SECs) contain two different transcription elongation factors, P-TEFb and ELL1/2, linked by the scaffolding protein AFF4 or AFF1. They stimulate the expression of both normal and disease-related genes, especially those of HIV or those involved in leukemogenesis. Among all SEC subunits, ELL2 is stoichiometrically limiting and uniquely regulated at the level of protein stability. Here we identify the RING domain protein Siah1, but not the homologous Siah2, as the E3 ubiquitin ligase for ELL2 polyubiquitination and proteasomal degradation. Siah1 cannot access and ubiquitinate ELL2 bound to AFF4, although, at high concentrations, it also degrades AFF4/1 to destroy SECs. Prostratin and HMBA, two well-studied activators of HIV transcription and latency, enhance ELL2 accumulation and SECs formation largely through decreasing Siah1 expression and ELL2 polyubiquitination. Given its importance in formation of SECs, the Siah1 ubiquitination pathway provides a fresh avenue for developing strategies to control disease-related transcription.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
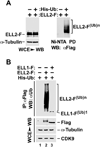
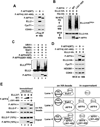
Figure 1. ELL2 but not the homologous ELL1 is a polyubiquitinated protein
A. HeLa cells were transfected with constructs expressing Flag-tagged ELL2 (ELL2-F) or/and Histidine-tagged ubiquitin (His-Ub) as indicated. Whole cell extracts (WCE) were prepared and subjected to analysis by Western blotting (WB) for the presence of the indicated proteins (left panel) or precipitation with Ni2+-NTA beads to pull down (PD) His-Ub, whose covalently bound ELL2-F was detected by anti-Flag WB (right panel). B. WCE were prepared from HeLa cells transfected with the indicated expression constructs and analyzed by WB directly (bottom panels) or anti-Flag immunoprecipitation (IP), which was followed by WB with anti-Ub antibody (top panel). ELL2-F(Ub)n and ELL1-F(Ub)1 indicate polyubiquitinated ELL2 and monoubiquitinated ELL1, respectively.
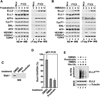
Figure 2. Siah1 depletion suppresses ELL2 polyubiquitination and promotes ELL2 stability, SEC formation and SEC-dependent HIV-1 transcription
A. The HA-Siah1 expression plasmid was co-transfected into HeLa cells with an empty vector (ctrl) or plasmids expressing the indicated shRNAs targeting Siah1. HA-Siah1 and α-tubulin present in cell lysates were examined by Western blotting. B. Whole cell lysates (WCE) of cells expressing the indicated shSiah1 sequences were analyzed by Western Blotting (WB) for the indicated proteins. C. WCE examined in B were subjected to immunoprecipitation with either anti-CDK9 Ab or total rabbit (ctl.) IgG and immunoprecipitates were analyzed by WB for the indicated proteins. D. Polyubiquitinated ELL2-F(Ub)n isolated by pull-down with Ni2+-NTA beads and total ELL2-F in WCE of cells expressing the indicated shSiah1 sequences were examined by anti-Flag WB. E. Luciferase activities were measured in extracts of cells co-transfected with the indicated shSiah1-expressing constructs, the HIV-1 LTR-luciferase reporter gene, and a vector expressing Tat or nothing. The activity in cells expressing the non-effective shSiah1 #3 but not Tat was set to 1. The error bars represent mean +/− SD from three independent sets of transfection.
Figure 3. ELL2 is more susceptible than AFF1 and AFF4 to degradation induced by Siah1, an E3 ubiquitin ligase that directly polyubiquitinates the ELL2 C-terminal region in vivo and in vitro
A. & B. HeLa cells were transfected with (+) or without (−) the indicated expression constructs and whole cell lysates (WCE) were examined by Western blotting (WB) with the indicated antibodies. The HA-Siah1 mRNA levels produced from the transfected plasmid were determined by qRT-PCR and the ratios over endogenous GAPDH mRNA were shown in the bottom panel of A. The error bars represent mean +/− SD from three independent experiments. UD: undetectable. The HA-Siah1 construct was transfected in 3-fold increment in B. C. HeLa cells were transfected with plasmids expressing either WT or the C75S mutant HA-Siah1 and then treated with (+) or without (−) MG132. WCE were examined by WB for the indicated proteins. D. WCE of transfected cells were analyzed by WB as in A. E. WCE of HeLa cells transfected with the indicated expression constructs and anti-HA IP derived from WCE were analyzed by anti-HA and –Flag Western blotting. F. HeLa cells were transfected with constructs expressing WT or truncated ELL2-F and His-Ub as indicated. WCE as well as polyubiquitinated ELL2 isolated from WCE by Ni2+-NTA pull down (PD) were analyzed by anti-Flag WB as in Fig. 1A. G. In vitro ubiquitination reactions containing the indicated components were performed and 35S-labeled WT ELL2 was detected by autoradiography.
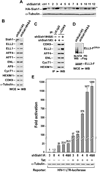
Figure 4. Sequence variations in the RING domains of Siah1 and Siah2 prevent the latter from inducing ELL2 degradation and inhibiting HIV transcription
A. HeLa cells were transfected with plasmids expressing either HA-Siah1 or HA-Siah2 and then treated with (+) or without (−) MG132. WCE were examined by WB for presence of the indicated proteins. B. Sequence alignment of the RING domains of Siah1 and Siah2, with the amino acid differences highlighted. C. WCE of cells co-transfected with the indicated plasmids were examined by WB for the indicated proteins. The HA-Siah1/2 constructs were introduced in 4-fold increment. D. Luciferase activities were measured in extracts of cells co-transfected with the HA-Siah1-expressing plasmid (in 3-fold increment), the HIV-1 LTR-luciferase reporter gene, and a vector expressing Tat or nothing. The activity in cells expressing neither HA-Siah1 nor Tat was set to 1. The error bars represent mean +/− SD from three independent experiments. E. Cells were transfected with the indicated WT or mutant HA-Siah2 constructs in 3-fold increment and luciferase activities were examined and analyzed as in D.
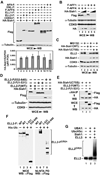
Figure 5. The AFF4-ELL2 interaction sequesters ELL2 away from Siah1 to inhibit Siah1-induced polyubiquitination and degradation of ELL2
A. Anti-Flag immunoprecipitates (IP) derived from nuclear extracts of HeLa cells transfectd with the indicated F-AFF4 expression plasmids were analyzed by Western blotting (WB) for presence of the indicated proteins. B. Cells were transfected with the various expression plasmids as indicated at the top. Whole cell extracts (WCE) as well as His-Ub and the covalently bound ELL2-HA isolated from WCE by Ni2+-NTA pull down (PD) were examined by WB for the indicated proteins. C. In vitro ubiquitination reactions were performed with the indicated components. The 35S-labeled ELL2 and F-AFF4 proteins were detected by autoradiography and anti-Flag Western blotting, respectively. D. Cells were co-transfected with a constant amount of the plasmid producing ELL2-HA and the plasmid expressing WT F-AFF4, the Δ301–400 mutant or nothing. WCE were prepared and analyzed by WB for the indicated proteins. E. In vitro binding reactions contained constant amounts of HA-Siah1 immobilized on anti-HA beads and ELL2-F in solution. WT or Δ301–400 F-AFF4 were either not added (−) or added (+) in 3-fold increment into the binding reactions. The bound and input proteins were examined by Western blotting as indicated. The diagram on the right depicts the likely scenarios as encountered in the indicated reactions.
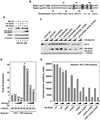
Figure 6. Activators of HIV transcription and latency promote ELL2 accumulation and SECs formation through inhibiting Siah1 expression and ELL2 polyubiquitination
A. & B. F1C2 cells stably expressing CDK9-F were treated with prostratin (A) or HMBA (B) for the indicated periods of time. Nuclear extracts (NE) and anti-Flag immunoprecipitates (IP) derived from NEs were analyzed by Western blotting (WB) for the indicated proteins. C. Siah1 protein isolated from HeLa cells treated with the indicated compounds was analyzed by WB. D. The Siah1 mRNA levels relative to those of GAPDH were determined by qRT-PCR, and the ratio obtained in cells treated with DMSO was set to 100%. The error bars represent mean +/− SD from three independent experiments. E. HeLa cells expressing His-Ub or not were treated with the indicated chemicals for 6 hrs. Whole cell extracts (WCE) as well as His-Ub and the covalently bound ELL2 isolated by Ni2+-NTA pull down (PD) were analyzed by WB for the indicated proteins.
Similar articles
- The PARP1-Siah1 Axis Controls HIV-1 Transcription and Expression of Siah1 Substrates.
Yu D, Liu R, Yang G, Zhou Q. Yu D, et al. Cell Rep. 2018 Jun 26;23(13):3741-3749. doi: 10.1016/j.celrep.2018.05.084. Cell Rep. 2018. PMID: 29949759 Free PMC article. - Host cell factors stimulate HIV-1 transcription by antagonizing substrate-binding function of Siah1 ubiquitin ligase to stabilize transcription elongation factor ELL2.
Wu J, Xue Y, Gao X, Zhou Q. Wu J, et al. Nucleic Acids Res. 2020 Jul 27;48(13):7321-7332. doi: 10.1093/nar/gkaa461. Nucleic Acids Res. 2020. PMID: 32479599 Free PMC article. - Gene target specificity of the Super Elongation Complex (SEC) family: how HIV-1 Tat employs selected SEC members to activate viral transcription.
Lu H, Li Z, Zhang W, Schulze-Gahmen U, Xue Y, Zhou Q. Lu H, et al. Nucleic Acids Res. 2015 Jul 13;43(12):5868-79. doi: 10.1093/nar/gkv541. Epub 2015 May 24. Nucleic Acids Res. 2015. PMID: 26007649 Free PMC article. - The inducible E3 ubiquitin ligases SIAH1 and SIAH2 perform critical roles in breast and prostate cancers.
Knauer SK, Mahendrarajah N, Roos WP, Krämer OH. Knauer SK, et al. Cytokine Growth Factor Rev. 2015 Aug;26(4):405-13. doi: 10.1016/j.cytogfr.2015.04.002. Epub 2015 May 12. Cytokine Growth Factor Rev. 2015. PMID: 26028498 Review. - Siah1 in cancer and nervous system diseases (Review).
Zhang H, Wang J, Ge Y, Ye M, Jin X. Zhang H, et al. Oncol Rep. 2022 Feb;47(2):35. doi: 10.3892/or.2021.8246. Epub 2021 Dec 27. Oncol Rep. 2022. PMID: 34958110 Review.
Cited by
- Regulation of ELL2 stability and polyubiquitination by EAF2 in prostate cancer cells.
Yang T, Jing Y, Dong J, Yu X, Zhong M, Pascal LE, Wang D, Zhang Z, Qiao B, Wang Z. Yang T, et al. Prostate. 2018 Nov;78(15):1201-1212. doi: 10.1002/pros.23695. Epub 2018 Jul 15. Prostate. 2018. PMID: 30009504 Free PMC article. - Pirin down-regulates the EAF2/U19 protein and alleviates its growth inhibition in prostate cancer cells.
Qiao Z, Wang D, Hahn J, Ai J, Wang Z. Qiao Z, et al. Prostate. 2014 Feb;74(2):113-20. doi: 10.1002/pros.22729. Epub 2013 Nov 23. Prostate. 2014. PMID: 24272884 Free PMC article. - The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat.
Schulze-Gahmen U, Upton H, Birnberg A, Bao K, Chou S, Krogan NJ, Zhou Q, Alber T. Schulze-Gahmen U, et al. Elife. 2013 Mar 5;2:e00327. doi: 10.7554/eLife.00327. Elife. 2013. PMID: 23471103 Free PMC article. - Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure.
Mbonye U, Karn J. Mbonye U, et al. Virology. 2014 Apr;454-455:328-39. doi: 10.1016/j.virol.2014.02.008. Epub 2014 Feb 22. Virology. 2014. PMID: 24565118 Free PMC article. Review. - Dynamic Change of Transcription Pausing through Modulating NELF Protein Stability Regulates Granulocytic Differentiation.
Liu X, Gogate AA, Tastemel M, Malladi VS, Yao H, Nguyen K, Huang LJ, Bai X. Liu X, et al. Blood Adv. 2017 Aug 8;1(18):1358-1367. doi: 10.1182/bloodadvances.2017008383. Blood Adv. 2017. PMID: 28868519 Free PMC article.
References
- Apcher GS, Heink S, Zantopf D, Kloetzel PM, Schmid HP, Mayer RJ, Kruger E. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal alpha and beta subunits. FEBS letters. 2003;553:200–204. - PubMed
- Bursen A, Moritz S, Gaussmann A, Dingermann T, Marschalek R. Interaction of AF4 wild-type and AF4.MLL fusion protein with SIAH proteins: indication for t(4;11) pathobiology? Oncogene. 2004;23:6237–6249. - PubMed
- Carthew RW, Rubin GM. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. - PubMed
- Chin LS, Vavalle JP, Li L. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem. 2002;277:35071–35079. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AI095057/AI/NIAID NIH HHS/United States
- R01 AI095057/AI/NIAID NIH HHS/United States
- R01 AI041757/AI/NIAID NIH HHS/United States
- R01AI41757/AI/NIAID NIH HHS/United States
- F31 GM082156/GM/NIGMS NIH HHS/United States
- F31GM082156/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases