Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor - PubMed (original) (raw)
. 2012 Apr 20;36(4):561-71.
doi: 10.1016/j.immuni.2012.02.014. Epub 2012 Apr 5.
Andrew Perry, Jiansheng Jiang, Patrick Smith, James A Curry, Leonie Unterholzner, Zhaozhao Jiang, Gabor Horvath, Vijay A Rathinam, Ricky W Johnstone, Veit Hornung, Eicke Latz, Andrew G Bowie, Katherine A Fitzgerald, T Sam Xiao
Affiliations
- PMID: 22483801
- PMCID: PMC3334467
- DOI: 10.1016/j.immuni.2012.02.014
Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor
Tengchuan Jin et al. Immunity. 2012.
Abstract
Recognition of DNA by the innate immune system is central to antiviral and antibacterial defenses, as well as an important contributor to autoimmune diseases involving self DNA. AIM2 (absent in melanoma 2) and IFI16 (interferon-inducible protein 16) have been identified as DNA receptors that induce inflammasome formation and interferon production, respectively. Here we present the crystal structures of their HIN domains in complex with double-stranded (ds) DNA. Non-sequence-specific DNA recognition is accomplished through electrostatic attraction between the positively charged HIN domain residues and the dsDNA sugar-phosphate backbone. An intramolecular complex of the AIM2 Pyrin and HIN domains in an autoinhibited state is liberated by DNA binding, which may facilitate the assembly of inflammasomes along the DNA staircase. These findings provide mechanistic insights into dsDNA as the activation trigger and oligomerization platform for the assembly of large innate signaling complexes such as the inflammasomes.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1. The HIN:DNA Interactions are Non-sequence Specific and Sensitive to Ionic Strength
(A) Fluorescence polarization (FP) assays of the FAM-labeled dsDNAs of various sequences and GC contents upon binding to the AIM2 HIN domain. The apparent Kd values (Kda) are shown for each dsDNA. (B) FP assays of the FAM-labeled dsDNA ODN787/788 upon binding to the AIM2 HIN domain in the presence of various concentrations of sodium chloride. (C) FP assays of the FAM-labeled dsDNA ODN787/788 upon binding to the IFI16 HINb domain in the presence of various concentrations of sodium chloride.
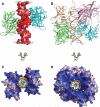
Figure 2. Overview of The HIN:DNA Complexes
(A) The structure of the AIM2 HIN:DNA complex (crystal form I) is represented as lime and cyan-colored ribbons for each HIN domain and electrostatic charge surface for the dsDNA on a scale of -10 kT/e (red) to 10 kT/e (blue). Locations of the N-termini of the HIN domains are marked. (B) Structure of the IFI16 HINb:DNA complex is shown as lime, cyan, lightpink and wheat ribbons for each HINb domain and orange ribbon for the dsDNA. Locations of the N-termini of the HIN domains are marked. (C) Structure of the AIM2 HIN:DNA complex is rotated 90 degrees along the horizontal axis from the view in (A), and represented as electrostatic charge surface for the HIN domains and orange ribbon for the dsDNA. (D) Structure of the IFI16 HINb:DNA complex is represented as electrostatic charge surface for the HIN domains and orange ribbon for the dsDNA. The view is rotated 90 degrees horizontally from that in (B). See also Figure S1.
Figure 3. The HIN Domains Bind Both Strands of The dsDNA
(A) Detailed HIN:DNA interactions for the AIM2 HIN:DNA complex. The hydrogen bonds are indicated as gray dotted lines. Secondary structures for the AIM2 HIN domain (lime) are labeled and the two DNA strands are colored yellow and silver, respectively. The approximate boundaries of the OB1-OB2 are marked with a magenta dotted line and the major and minor grooves of the dsDNA are marked in gray. (B) Detailed HIN:DNA interactions are shown for the IFI16 HINb:DNA complex similar to (A), except the IFI16 HINb domain is colored cyan. (C) Sequence alignment of the HIN domains. Sequences of selected dsDNA-binding HIN domains from human AIM2 (NP_004824), mouse AIM2 (NP_001013801), human IFI16 (Q16666), mouse p204 (NP_032355, a homolog of human IFI16), mouse p202 (NP_032353, an inhibitor of AIM2), as well as a ssDNA-binding OB superfamily protein RPA (NP_002936) were aligned by ClustalW (Thompson et al., 1994) with minor adjustments. The α helices are in red, and the β strands were underlined in green and marked with “I” and “II” for OB1 and OB2, respectively. Conserved residues are shaded in yellow, and DNA binding residues in black boxes. See also Figure S2.
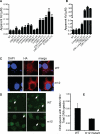
Figure 4. Mutagenesis Studies of The HIN:DNA Interactions
(A) Wild type and mutant AIM2 HIN domains were assayed for DNA binding and their apparent Kd values are plotted in a bar graph. Locations of the mutations for each mutant are marked below the graph. The “m0” mutants contain mutations of basic residues outside the DNA-binding surface. More details on the mutation sites are listed in Table S4. Panels of the binding curves for each mutant are presented in Figure S3A. (B) Wild type and mutant IFI16 HINb domains were assayed for DNA binding and their apparent Kd values are plotted in a bar graph. Locations of the mutations for each mutant are marked below the graph. The “m0” mutants contain mutations of basic residues outside the DNA-binding surface. More details on the mutation sites are listed in Table S4. Panels of the binding curves for each mutant are presented in Figure S3D. (C) Co-localization of DNA with wild type or mutant (m12) AIM2 HIN domains containing a hemagglutinin (HA) tag in HEK293T cells. Cells were stained 24 hours after transfection for AIM2 (anti-HA antibody with Alexa647, red) and DNA (DAPI, blue). Regions of colocalization are marked with white arrows. (D) Co-localization of FITC-labeled dsDNA (red) with stably expressed wild type or mutant (m12) AIM2 HIN domains tagged with mCerulean (green) in AIM2 deficient macrophages. Regions of colocalization are marked with white arrows. (E) The ratio of the number of dsDNA specks that colocalized with the wild type or mutant AIM2 HIN domains to the total number of dsDNA specks from (D) are shown for 2 full 1200×1200 pixel mosaic images, respectively. See also Figure S3.
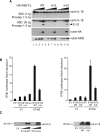
Figure 5. Innate immune Responses by The Full-length AIM2 and IFI16 Receptors
(A) Reconstitution of the human AIM2 inflammasome using the wild type, or mutant (m12 and m10) full-length HA-AIM2 and ASC, procaspase-1 and luciferase-FLAG tagged pro-IL-1β. Maturation of the IL-1β is indicated with a black arrow. The expression levels of the wild type or mutant full-length HA-AIM2 are indicated at the bottom panels as probed by anti-HA or anti-AIM2 antibodies. (B) Interferon-β promoter reporter assay for the wild type full-length IFI16 or IFI16 containing the HINb mutant m4 or m5 in HEK293T cells. EV indicates empty expression vector. (C) The expression levels of the wild type or mutant full-length IFI16 proteins in (B) are detected by immunoblotting 48 hours after transfection.
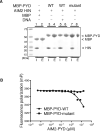
Figure 6. PYD domain Inhibits The HIN:DNA Interaction
(A) Pull-down assay of the MBP, MBP-PYD wild type or mutant (containing mutations of E7A, L11A, D15A, D19A and E20A) with the AIM2 HIN domain. The wild type MBP-PYD (lane 4), but not mutant MBP-PYD (lane 8) or MBP (lane 2), was able to pull down the AIM2 HIN domain, which was significantly reduced by the addition of a 19mer dsDNA ODN 736/737 (lane 6). “I”: input; “E”: elution. (B) The AIM2 PYD domain inhibits the HIN:DNA interaction. Increasing concentrations of the wild type or mutant AIM2 PYD domain were incubated with an AIM2 HIN:DNA mixture, and the fluorescence polarization was measured and analyzed with program Prism. The IC50 for the wild type AIM2 PYD domain is 11 μM. See also Figure S4.
Figure 7. Secretion of IL-1β induced by DNA of various lengths
Human PBMCs were primed with LPS and transfected with the indicated dsDNA, or MSU and ATP as controls. The culture supernatants were assayed for IL-1β secretion 6 hours after transfection/stimulation. See also Figure S5.
Similar articles
- Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling.
Li H, Wang J, Wang J, Cao LS, Wang ZX, Wu JW. Li H, et al. Acta Crystallogr F Struct Biol Commun. 2014 Jan;70(Pt 1):21-9. doi: 10.1107/S2053230X1303135X. Epub 2013 Dec 24. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 24419611 Free PMC article. - Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly.
Jin T, Perry A, Smith P, Jiang J, Xiao TS. Jin T, et al. J Biol Chem. 2013 May 10;288(19):13225-35. doi: 10.1074/jbc.M113.468033. Epub 2013 Mar 25. J Biol Chem. 2013. PMID: 23530044 Free PMC article. - The emerging role of human PYHIN proteins in innate immunity: implications for health and disease.
Connolly DJ, Bowie AG. Connolly DJ, et al. Biochem Pharmacol. 2014 Dec 1;92(3):405-14. doi: 10.1016/j.bcp.2014.08.031. Epub 2014 Sep 6. Biochem Pharmacol. 2014. PMID: 25199457 - The nucleic acid-sensing inflammasomes.
Xiao TS. Xiao TS. Immunol Rev. 2015 May;265(1):103-11. doi: 10.1111/imr.12281. Immunol Rev. 2015. PMID: 25879287 Free PMC article. Review. - Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization.
Veeranki S, Choubey D. Veeranki S, et al. Mol Immunol. 2012 Jan;49(4):567-71. doi: 10.1016/j.molimm.2011.11.004. Epub 2011 Dec 2. Mol Immunol. 2012. PMID: 22137500 Free PMC article. Review.
Cited by
- Innate immune recognition of DNA: A recent history.
Dempsey A, Bowie AG. Dempsey A, et al. Virology. 2015 May;479-480:146-52. doi: 10.1016/j.virol.2015.03.013. Epub 2015 Mar 26. Virology. 2015. PMID: 25816762 Free PMC article. Review. - Atrial Fibrillation in Chronic Kidney Disease: An Overview.
Gadde S, Kalluru R, Cherukuri SP, Chikatimalla R, Dasaradhan T, Koneti J. Gadde S, et al. Cureus. 2022 Aug 7;14(8):e27753. doi: 10.7759/cureus.27753. eCollection 2022 Aug. Cureus. 2022. PMID: 36106212 Free PMC article. Review. - Molecular characterization of woodchuck IFI16 and AIM2 and their expression in woodchucks infected with woodchuck hepatitis virus (WHV).
Yan Q, Li M, Liu Q, Li F, Zhu B, Wang J, Lu Y, Liu J, Wu J, Zheng X, Lu M, Wang B, Yang D. Yan Q, et al. Sci Rep. 2016 Jun 29;6:28776. doi: 10.1038/srep28776. Sci Rep. 2016. PMID: 27354260 Free PMC article. - Kaposi's sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes.
Singh VV, Kerur N, Bottero V, Dutta S, Chakraborty S, Ansari MA, Paudel N, Chikoti L, Chandran B. Singh VV, et al. J Virol. 2013 Apr;87(8):4417-31. doi: 10.1128/JVI.03282-12. Epub 2013 Feb 6. J Virol. 2013. PMID: 23388709 Free PMC article. - Structural analysis of the HIN1 domain of interferon-inducible protein 204.
Tian Y, Yin Q. Tian Y, et al. Acta Crystallogr F Struct Biol Commun. 2019 Jun 1;75(Pt 6):455-460. doi: 10.1107/S2053230X19007167. Epub 2019 Jun 10. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 31204693 Free PMC article.
References
- Aksentijevich I, Kastner DL. Genetics of monogenic autoinflammatory diseases: past successes, future challenges. Nat Rev Rheumatol. 2011;7:469–478. - PubMed
- Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ZIA AI000960-06/ImNIH/Intramural NIH HHS/United States
- R37 AI067497/AI/NIAID NIH HHS/United States
- R01 AI067497/AI/NIAID NIH HHS/United States
- R56 AI067497/AI/NIAID NIH HHS/United States
- AI067497/AI/NIAID NIH HHS/United States
- R01 AI093752/AI/NIAID NIH HHS/United States
- 243046/ERC_/European Research Council/International
- R01 AI083713/AI/NIAID NIH HHS/United States
- AI083713/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases