Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575 - PubMed (original) (raw)
Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575
Sabine Figaro et al. Mol Cell Biol. 2012 Jun.
Abstract
Posttranscriptional and posttranslational modification of macromolecules is known to fine-tune their functions. Trm112 is unique, acting as an activator of both tRNA and protein methyltransferases. Here we report that in Saccharomyces cerevisiae, Trm112 is required for efficient ribosome synthesis and progression through mitosis. Trm112 copurifies with pre-rRNAs and with multiple ribosome synthesis trans-acting factors, including the 18S rRNA methyltransferase Bud23. Consistent with the known mechanisms of activation of methyltransferases by Trm112, we found that Trm112 interacts directly with Bud23 in vitro and that it is required for its stability in vivo. Consequently, trm112Δ cells are deficient for Bud23-mediated 18S rRNA methylation at position G1575 and for small ribosome subunit formation. Bud23 failure to bind nascent preribosomes activates a nucleolar surveillance pathway involving the TRAMP complexes, leading to preribosome degradation. Trm112 is thus active in rRNA, tRNA, and translation factor modification, ideally placing it at the interface between ribosome synthesis and function.
Figures
Fig 1
Trm112 is required for small ribosomal subunit accumulation, efficient preribosome export of both ribosomal subunits, and progression through mitosis. (A) (Left) _trm112_Δ cells are hypersensitive to paromomycin. Serial dilutions of _trm112_Δ (yVH90) yeast cells and the isogenic wild type (WT) were spotted on complete medium (YPDA), supplemented or not with paromomycin (800 μg/ml). (Right) _trm112_Δ cells are cryosensitive for growth, and the TRM112-TAP allele is fully functional (no growth defect). Cells were grown for 2 days at 30°C and 37°C and for 3 days at 23°C. (B) _trm112_Δ cells are characterized by a ribosomal subunit imbalance. Shown is a polysome profile (absorbance at 254 nm) analysis of a _trm112_Δ strain and the wild-type isogenic control. The positions of 40S, 60S, 80S, and polysomes are indicated. (C) The amount of both the small and the large ribosomal subunits is reduced in _trm112_Δ cells. Analysis was conducted under dissociation conditions (absence of MgCl2) for determination of the amount of small and large ribosomal subunits. This experiment was repeated 4 times. (D) Preribosome export assay. _trm112_Δ cells and the isogenic wild type (BY4741) were transformed with a construct expressing either Rps2-GFP or Rpl25-GFP, grown to mid-log phase, and analyzed by fluorescence under a microscope. The arrow points to a nucleolar focus. Arrowheads point to nucleolar foci asymmetrically localized in the mother cell. The insets highlight cells blocked in mitosis with an asymmetrically localized nucleolar focus. DIC, differential interference contrast; DAPI, 4′,6-diamidino-2-phenylindole.
Fig 2
Trm112 is required for efficient pre-rRNA processing, and the absence of Trm112 activates a 3′→5′ nucleolar surveillance pathway which targets preribosomes for degradation. (A) In _trm112_Δ cells, early pre-rRNA processing is inhibited and precursors to large subunit rRNA are unstable. (Left) Schematics depicting the pre-rRNA intermediates and probes used. (Right) Total RNA extracted from the indicated strains grown to mid-log phase in YPDA at 30°C (gels I to VII) or 23°C (gels VIII and IX) was separated on agarose-formaldehyde gels, transferred to a nylon membrane, and hybridized with a set of oligonucleotide probes in Northern blots, as indicated on the gels. Gel VI was hybridized with LD915 and gel VII with LD1290. (B) In _trm112_Δ cells, pre-rRNA precursors are turned over by TRAMP-dependent nucleolar surveillance. Information is the same as for panel A. Cells were grown at 30°C. Gel I, ethidium bromide staining.
Fig 3
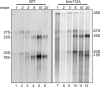
Trm112 is required for efficient small ribosomal subunit rRNA processing. _trm112_Δ and isogenic wild-type (WT) cells, transformed with a plasmid expressing the URA3 gene, were grown at 23°C in minimal medium lacking uracil, pulse-labeled with tritiated uracil for 1 min, and chased with an excess of cold uracil for the indicated times. Total RNA was extracted, separated on denaturing agarose gels, and transferred to a GeneScreen membrane. The membrane was sprayed with tritium enhancer and exposed to film. Lanes corresponding to _trm112_Δ cells were exposed 4 times longer than those of the WT control.
Fig 4
Trm112 copurifies with preribosomes that also contain the 18S rRNA methyltransferase Bud23. Partners of Trm112 were identified by affinity purification followed by mass spectrometry analysis. Yeast cells expressing a functional Trm112-TAP fusion (see Fig. 1A) were grown exponentially to mid-log phase at 30°C in YPDA. Total lysates were prepared in solid phase (see Materials and Methods) and engaged in a revised affinity purification protocol (40). As a control, we used a strain expressing the TAP tag alone (data not shown). Interactions were carefully curated with those identified in the TAP-alone control and with an unrelated bait. All interactions are listed in Table S3 in the supplemental material. Interactants were identified by LC-MS/MS. (Left) Major Trm112 interactants sorted and color-coded by known or putative functions. RPS and RPL, ribosomal proteins from the small and large subunit, respectively. (Right) Eluate from a representative Trm112-TAP purification analyzed by 4 to 12% NuPage Novex bis-tris (Invitrogen) followed by Coomassie blue staining. Trm112-TAP is highlighted with an asterisk.
Fig 5
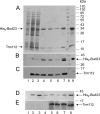
Trm112 interacts with pre-90S and pre-40S ribosomes. (A) Velocity gradient analysis. A total protein extract from wild-type cells was fractionated on a 10 to 50% sucrose gradient. A total of 24 fractions were collected and analyzed by Western blotting with an antibody raised against the recombinant Bud23-Trm112 complex or with antibodies specific to Nog1 or ribosomal proteins S8 and L3. (B and C) Primer extension analysis of RNA coprecipitating with Trm112. Total extracts from yeast cells expressing Trm112-TAP, Nop58-TAP, Enp1-TAP, or TAP alone were engaged in affinity purification. Coprecipitating RNAs were analyzed by primer extension with oligonucleotide LD2484 (B) or LD471 (C). (B) Lanes 1 to 4, pellets from the indicated strains; lane 5, total RNA from Trm112-TAP. The total and pellet fractions were loaded in a 1:9 ratio. (C) Lanes 1, 2, 4, and 5, pellets from the indicated strains; lanes 3, total RNA from cells expressing the TAP alone; lane 6, total RNA from Trm112-TAP.
Fig 6
Trm112 interacts directly with the 18S rRNA methyltransferase Bud23 in vitro, and Trm112 increases the solubility of Bud23. (A) Coomassie blue staining of an SDS-PAGE gel showing coelution with imidazole of a poly-His-tagged Bud23 with untagged Trm112 from a nickel affinity matrix. Lane 1, uninduced culture; lane 2, total bacterial extract from cells coexpressing His6-Bud23 and untagged Trm112, respectively, from pACDuet(His6Bud23) and pET11a(Trm112); lane 3, flowthrough; lanes 4 to 7, coelution in a 50 mM imidazole-containing buffer (20 μl of eluted fractions in each lane); lane 8, molecular mass marker; lane 9, 10 μl of purified His6-Bud23-Trm112 complex following a 5× concentration through Amicon 30 (Millipore). (B) Western blot identification of His6-Bud23. The SDS-PAGE gel shown in panel A was duplicated, transferred to nitrocellulose, and probed in a Western blot with an anti-His6 antibody. (C) Information same as for panel B. Trm112 was detected with an antibody specific to the Mtq2-Trm112 complex (that only reveals Trm112 here since these cells do not express Mtq2). (D and E) The coexpression of Trm112 and Bud23 increases the solubility of recombinant Bud23. Lanes 1 to 4, E. coli cells expressing only His6-Bud23; lanes 5 to 7, E. coli cells coexpressing His6-Bud23 and Trm112; lane 8, control purified His6-Bud23-Trm112 complex. Lane 1 contains extract from noninduced cells, lanes 2 and 5 contain total extract from induced cells, lanes 3 and 6 contain pellets from induced cells, and lanes 4 and 7 contain supernatant from induced cells. (D) Western blot detection of His6-Bud23 with an anti-His antibody. (E) Western blot detection of Trm112 with a specific antibody.
Fig 7
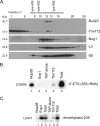
Trm112 is required for Bud23-mediated formation of the _N_7-methylguanosine at position G1575 in 18S rRNA. Total RNA extracted from _bud23_Δ and _trm112_Δ cells and an isogenic wild-type strain (WT, BY4741) grown to mid-log phase at 30°C in YPDA was analyzed by primer extension from oligonucleotide LD2092, hybridizing 140 nucleotides downstream of the site of modification. (A) Reverse transcription on total RNA. The reverse transcriptase is blocked at the site of modification G1575. The G1575 modification is best detected at the low dNTP concentration of 0.04 mM, which was used here. (B) Reverse transcription on RNAs that have specifically been cleaved at N7-methylated guanosine positions. The reverse transcriptase stops at the site of RNA cleavage, revealing position G1575. The control consisted of untreated RNAs; in the case of chemically treated RNA, total RNA was treated with NaBH4 and aniline to break the phosphodiester backbone at the m7G position (see Materials and Methods), and dimethyl sulfate-modified tRNA was added in the reaction to enhance cleavage at m7G methylation sites. In panel B, the regular dNTP concentration of 1.20 mM was used. Note that by comparison to panel A, which used the lower dNTP concentration of 0.04 mM, a stronger extension ladder was seen in the background in panel B. In panels A and B, the asterisk denotes a structural stop in the substrate and indicates that the same amount of reverse-transcribed RNA was loaded in each lane. In both panels A and B, a 2′-O methylation at position G1572, directed by snoRNA57, was detected 3 nucleotides above G1575 (discussed in reference 64). The detection of this modification was not affected by deletion of either BUD23 or TRM112. (C) Trm112 is required for Bud23 accumulation in vivo. Total protein was extracted from the strain indicated grown to mid-log phase in complete medium. Proteins were analyzed by SDS-PAGE and Western blotting with an antibody specific to the Bud23-Trm112 complex. Lane 1, 100 ng of recombinant His6-Bud23-Trm112 complex purified from E coli was loaded as a control; lanes 2 to 4, 50 μg of total protein. The asterisk denotes a cross-hybridizing band and provides a loading control.
Fig 8
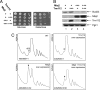
Several RNA and protein methyltransferases compete to interact with their cofactor Trm112.(A) Growth plate assay. Serial dilutions (10×) of wild-type cells (BY4741) overexpressing Mtq2, Trm112, or both from pGAL inducible promoters, and an isogenic control, were grown on glucose or galactose-based medium at 30°C for 2 days. +++, overexpression. (B) Western blot analysis of protein steady-state accumulation in strains presented in panel A. Bud23 and Mtq2-Trm112 were detected, respectively, with an antibody raised against the Bud23-Trm112 and Mtq2-Trm112 complexes (see Materials and Methods). Pgk1 (loading control) was detected with a specific commercial antibody. The overexpression of Mtq2 leads to an ∼5-fold reduction in the steady-state accumulation of Bud23 (average calculated on three independent experiments; SD, 0.7). (C) Polysome analysis (254-nm absorbance readings) of strains described for panel A. The arrow points to the 60S peak. The 40S/60S ratio in each panel was determined under dissociation conditions (see Fig. 1C). This experiment was repeated twice. WT, wild type.
Fig 9
Trm112 interaction with MTases. (A) Ribbon representation of the E. cuniculi Mtq2 (pink)-Trm112 complex. The Trm112 zinc-binding and central domains are shown in green and brown, respectively. The zinc atom is shown as a gray sphere. The SAM cofactor is depicted in sticks and its methyl group by a purple sphere. Inset to the right, enlarged view of the structural elements at the interface. (B) Ribbon representation of the S. cerevisiae Bud23 (orange)-Trm112 complex model. The Trm112 color code and orientation are the same as in panel A. The secondary structure elements predicted for Bud23 are indicated. (C) Overlay of the Mtq2-Trm112 complex structure (gray) and Bud23-Trm112 complex model (same color code as in panel B).
Similar articles
- Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes.
Létoquart J, Huvelle E, Wacheul L, Bourgeois G, Zorbas C, Graille M, Heurgué-Hamard V, Lafontaine DL. Létoquart J, et al. Proc Natl Acad Sci U S A. 2014 Dec 23;111(51):E5518-26. doi: 10.1073/pnas.1413089111. Epub 2014 Dec 8. Proc Natl Acad Sci U S A. 2014. PMID: 25489090 Free PMC article. - Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits.
White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. White J, et al. Mol Cell Biol. 2008 May;28(10):3151-61. doi: 10.1128/MCB.01674-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332120 Free PMC article. - The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits.
Sardana R, Johnson AW. Sardana R, et al. Mol Biol Cell. 2012 Nov;23(21):4313-22. doi: 10.1091/mbc.E12-05-0370. Epub 2012 Sep 5. Mol Biol Cell. 2012. PMID: 22956767 Free PMC article. - Trm112, a Protein Activator of Methyltransferases Modifying Actors of the Eukaryotic Translational Apparatus.
Bourgeois G, Létoquart J, van Tran N, Graille M. Bourgeois G, et al. Biomolecules. 2017 Jan 27;7(1):7. doi: 10.3390/biom7010007. Biomolecules. 2017. PMID: 28134793 Free PMC article. Review. - [The Common Partner of Several Methyltransferases Modifying the Components of The Eukaryotic Translation Apparatus].
Vasilieva EN, Laptev IG, Sergiev PV, Dontsova OA. Vasilieva EN, et al. Mol Biol (Mosk). 2018 Nov-Dec;52(6):975-983. doi: 10.1134/S0026898418060174. Mol Biol (Mosk). 2018. PMID: 30633240 Review. Russian.
Cited by
- The Stability of Ribosome Biogenesis Factor WBSCR22 Is Regulated by Interaction with TRMT112 via Ubiquitin-Proteasome Pathway.
Õunap K, Leetsi L, Matsoo M, Kurg R. Õunap K, et al. PLoS One. 2015 Jul 27;10(7):e0133841. doi: 10.1371/journal.pone.0133841. eCollection 2015. PLoS One. 2015. PMID: 26214185 Free PMC article. - Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1.
Sharma S, Langhendries JL, Watzinger P, Kötter P, Entian KD, Lafontaine DL. Sharma S, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2242-58. doi: 10.1093/nar/gkv075. Epub 2015 Feb 4. Nucleic Acids Res. 2015. PMID: 25653167 Free PMC article. - Dynamic methylome of internal mRNA N7-methylguanosine and its regulatory role in translation.
Malbec L, Zhang T, Chen YS, Zhang Y, Sun BF, Shi BY, Zhao YL, Yang Y, Yang YG. Malbec L, et al. Cell Res. 2019 Nov;29(11):927-941. doi: 10.1038/s41422-019-0230-z. Epub 2019 Sep 13. Cell Res. 2019. PMID: 31520064 Free PMC article. - N 2-methylguanosine modifications on human tRNAs and snRNA U6 are important for cell proliferation, protein translation and pre-mRNA splicing.
Wang C, Ulryck N, Herzel L, Pythoud N, Kleiber N, Guérineau V, Jactel V, Moritz C, Bohnsack MT, Carapito C, Touboul D, Bohnsack KE, Graille M. Wang C, et al. Nucleic Acids Res. 2023 Aug 11;51(14):7496-7519. doi: 10.1093/nar/gkad487. Nucleic Acids Res. 2023. PMID: 37283053 Free PMC article. - Identification of m7G-Related miRNA Signatures Associated with Prognosis, Oxidative Stress, and Immune Landscape in Lung Adenocarcinoma.
Jiang S, Xiao M, Shi Y, Wang Y, Xu Z, Wang K. Jiang S, et al. Biomedicines. 2023 May 29;11(6):1569. doi: 10.3390/biomedicines11061569. Biomedicines. 2023. PMID: 37371664 Free PMC article.
References
- Brunger AT. 2007. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2: 2728– 2733 - PubMed
- Chern MK, et al. 2009. Single-step protein purification by back flush in ion exchange chromatography. Anal. Biochem. 392: 174– 176 - PubMed
- Decatur WA, Fournier MJ. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27: 344– 351 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases