Immune response to bacteria induces dissemination of Ras-activated Drosophila hindgut cells - PubMed (original) (raw)
Immune response to bacteria induces dissemination of Ras-activated Drosophila hindgut cells
Erdem Bangi et al. EMBO Rep. 2012.
Abstract
Although pathogenic bacteria are suspected contributors to colorectal cancer progression, cancer-promoting bacteria and their mode of action remain largely unknown. Here we report that sustained infection with the human intestinal colonizer Pseudomonas aeruginosa synergizes with the Ras1V12 oncogene to induce basal invasion and dissemination of hindgut cells to distant sites. Cross-talk between infection and dissemination requires sustained activation by the bacteria of the Imd-dTab2-dTak1 innate immune pathway, which converges with Ras1V12 signalling on JNK pathway activation, culminating in extracellular matrix degradation. Hindgut, but not midgut, cells are amenable to this cooperative dissemination, which is progressive and genetically and pharmacologically inhibitable. Thus, Drosophila hindgut provides a valuable system for the study of intestinal malignancies.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
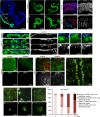
Ras1V12 expression is sufficient to initiate dissemination of the adult Drosophila hindgut cells. (A) Drosophila intestinal epithelium. Hindgut cells green fluorescent protein (GFP)-marked using byn-GAL4. Blue, nuclei (DAPI); m, midgut, r, rectum. (B–E) Hindguts expressing GFP alone (B,C) or GFP plus Ras1V12 (D,E) 7 days (B,D) or 21 days (C,E) after induction of transgenes. Arrowheads: cellular protrusions throughout ileum. (F,G) Surface views of ileum expressing GFP alone (F) or GFP plus Ras1V12 (G). Blue, laminin (basement membrane). (F′,G′) Laminin channel only of F and G. (H–K′) Cross-sections of control (H,H′) and Ras1V12 (I–K′) hindguts. Ras1V12-expressing hindguts (I–K) show GFP+ cells in the process of invading basally out of the gut epithelium (stars). These regions also have reduced or absent laminin staining (I′–K′, arrows). (L–N) Close-up views of disseminating cells (arrows) from Ras1V12-expressing hindguts. (laminin, blue). (L′,N′) Laminin channel only of L–N. Disseminating cells are present in regions of reduced laminin staining (arrows). (O–R) Matrix metalloprotease 1 (MMP1) expression (red) in control (O,P) and Ras1V12-expressing hindguts (Q,R). (O′–R′) MMP channel only of O–R. (S) Inside view of the abdominal cavity from an animal expressing Ras1V12 in the hindgut showing GFP+ foci (arrows). Stereoscope (T–Y) and confocal (W–Y) live views of GFP+ foci and dsRed nuclei (dsRed-nls). Note: yellow autoflorescence makes nephrocytes (n), fat body (f) and trachea (t) visible. (Z) Quantification of Ras1V12-induced dissemination over time. DAPI, 4,6-diamidino-2-phenylindole; nls, nuclear localization signal.
Figure 2
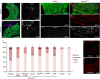
JNK pathway activation contributes to the induction of Ras1V12-mediated dissemination phenotypes. (A,B) PucZ expression in anterior (A) and posterior (B) byn>Ras1 V12 hindguts. (C,D) PucZ and matrix metalloprotease 1 (MMP1) expression in wild-type (C) and byn>Ras1 V12 (D) hindguts. A′,B′,C″,D″ are PucZ and C′,D′ are MMP1 channel only of A–D. (E) Quantitated dissemination 7 days after induction of the indicated transgenes (*P<0.05, **P_<0.01). (**F**,**G**) MMP1 of wild-type (**F**) and _byn>hep (G) hindguts. DAPI, 4,6-diamidino-2-phenylindole; GFP, green fluorescent protein.
Figure 3
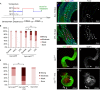
P. aeruginosa intestinal infection potentiates dissemination phenotypes. (A) Outline of the transgene induction and infection strategy. (B) Quantitated dissemination in flies fed on sucrose (suc) only, virulent (PA14) or avirulent (CF5) P. aeruginosa strain. (C,D) pucZ expression in infected (D) and uninfected (C) byn>Ras1 V12 hindguts. (C′,D′) pucZ only channels of C,D. (E) Quantitated dissemination in flies co-expressing Ras1V12 and Imd following the induction scheme in A without infection. (F, G) Hindguts co-expressing Ras1V12 and Imd show stronger MMP1 expression (red), particularly in the anterior region (arrows). (F′,G′) MMP1 channels of F,G (arrowheads: migratory cells; *P<0.05, **P<0.01). DAPI, 4,6-diamidino-2-phenylindole; GFP, green fluorescent protein; MMP1, matrix metalloprotease 1.
Figure 4
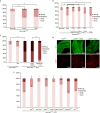
P. aeruginosa infection and Imd activation induce an earlier onset of dissemination phenotypes in the Ras1V12 background. (A) Quantitated dissemination 4 days after induction of Ras1V12 expression in the presence of infection. (B) CF5-induced dissemination in byn>Ras1V 12 flies co-expressing the indicated transgenes. (C) Co-expressing Imd, but not relD, with Ras1V12 results in a strong dissemination phenotype 4 days after induction. (D) MMP1 expression (red) in hindguts with indicated genotypes and treatment conditions 4 days after induction. Infection or Imd expression induces MMP1 expression in the presence of Ras1V12 4 days after induction. (E) Early onset of dissemination induced by infection is suppressed by RNAi and mutations of the indicated Imd pathway components. *P<0.05, **P<0.01. DAPI, 4,6-diamidino-2-phenylindole; DMSO, dimethylsulphoxide; GFP, green fluorescent protein; MMP1, matrix metalloprotease 1; RNAi, RNA interference.
Figure 5
Requirement for sustained bacterial infection for the induction and enhancement of dissemination. (A) Quantitated dissemination of byn>Ras1 V12 flies at 23 °C (semipermissive conditions) fed continuously on sucrose only or infected with CF5 for 4, 7 and 14 days (d). (B) Quantitated dissemination of byn>Ras1 V12 flies at days 7 and 14 of Ras1V12 transgene induction at 29 °C. Animals were either fed continuously on sucrose only (‘suc 7d’ and ‘suc 14d’) or infected with CF5 for 4 days followed by 3 or 10 days of bacterial clearance (‘CF5 4d + suc 3d’ or ‘CF5 4d + suc 10d’); or continuously infected for 7 or 14 days (‘CF5 7d’ and ‘CF5 14d’; **: P<0.01). (C) Model illustration depicting the synergistic induction of JNK and MMP1 in the hindgut enterocytes and the destruction of laminin as a mechanism that facilitates cell dissemination. (D) Model illustration delineating the branch of innate immune responses that converge with Ras1V12 signalling (red arrows) and the Imd pathway branch dedicated to the activation of Rel/nuclear factor-κB (black arrows). DAPI, 4,6-diamidino-2-phenylindole; DMSO, dimethylsulphoxide; GFP, green fluorescent protein; MMP1, matrix metalloprotease 1; NF-κB, nuclear factor-κB.
Similar articles
- Ras-oncogenic Drosophila hindgut but not midgut cells use an inflammation-like program to disseminate to distant sites.
Christofi T, Apidianakis Y. Christofi T, et al. Gut Microbes. 2013 Jan-Feb;4(1):54-9. doi: 10.4161/gmic.22429. Epub 2012 Oct 12. Gut Microbes. 2013. PMID: 23060054 Free PMC article. Review. - Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB.
Zhuang ZH, Sun L, Kong L, Hu JH, Yu MC, Reinach P, Zang JW, Ge BX. Zhuang ZH, et al. Cell Signal. 2006 Jul;18(7):964-70. doi: 10.1016/j.cellsig.2005.08.020. Epub 2005 Nov 28. Cell Signal. 2006. PMID: 16311020 - Synergy between bacterial infection and genetic predisposition in intestinal dysplasia.
Apidianakis Y, Pitsouli C, Perrimon N, Rahme L. Apidianakis Y, et al. Proc Natl Acad Sci U S A. 2009 Dec 8;106(49):20883-8. doi: 10.1073/pnas.0911797106. Epub 2009 Nov 23. Proc Natl Acad Sci U S A. 2009. PMID: 19934041 Free PMC article. - In Drosophila, RhoGEF2 cooperates with activated Ras in tumorigenesis through a pathway involving Rho1-Rok-Myosin-II and JNK signalling.
Khoo P, Allan K, Willoughby L, Brumby AM, Richardson HE. Khoo P, et al. Dis Model Mech. 2013 May;6(3):661-78. doi: 10.1242/dmm.010066. Epub 2013 Jan 11. Dis Model Mech. 2013. PMID: 23324326 Free PMC article. - Genetics of immune recognition and response in Drosophila host defense.
Ligoxygakis P. Ligoxygakis P. Adv Genet. 2013;83:71-97. doi: 10.1016/B978-0-12-407675-4.00002-X. Adv Genet. 2013. PMID: 23890212 Review.
Cited by
- Animal models to study acute and chronic intestinal inflammation in mammals.
Jiminez JA, Uwiera TC, Douglas Inglis G, Uwiera RR. Jiminez JA, et al. Gut Pathog. 2015 Nov 10;7:29. doi: 10.1186/s13099-015-0076-y. eCollection 2015. Gut Pathog. 2015. PMID: 26561503 Free PMC article. Review. - Intestinal Immune Deficiency and Juvenile Hormone Signaling Mediate a Metabolic Trade-off in Adult Drosophila Females.
Shianiou G, Teloni S, Apidianakis Y. Shianiou G, et al. Metabolites. 2023 Feb 24;13(3):340. doi: 10.3390/metabo13030340. Metabolites. 2023. PMID: 36984780 Free PMC article. - A feed-forward loop between Toll/NF-κB and Rac1 promotes epithelial to mesenchymal transition of Ras-oncogenic hindgut enterocytes in Drosophila.
Panagi M, Galaras A, Hatzis P, Apidianakis Y. Panagi M, et al. Biol Open. 2025 Jun 15;14(6):bio061960. doi: 10.1242/bio.061960. Epub 2025 Jun 17. Biol Open. 2025. PMID: 40476337 Free PMC article. - Functional analysis of PGRP-LA in Drosophila immunity.
Gendrin M, Zaidman-Rémy A, Broderick NA, Paredes J, Poidevin M, Roussel A, Lemaitre B. Gendrin M, et al. PLoS One. 2013 Jul 26;8(7):e69742. doi: 10.1371/journal.pone.0069742. Print 2013. PLoS One. 2013. PMID: 23922788 Free PMC article. - Ras-oncogenic Drosophila hindgut but not midgut cells use an inflammation-like program to disseminate to distant sites.
Christofi T, Apidianakis Y. Christofi T, et al. Gut Microbes. 2013 Jan-Feb;4(1):54-9. doi: 10.4161/gmic.22429. Epub 2012 Oct 12. Gut Microbes. 2013. PMID: 23060054 Free PMC article. Review.
References
- Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP, Raul F (2000) Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis 21: 753–756 - PubMed
- Guarino M, Rubino B, Ballabio G (2007) The role of epithelial-mesenchymal transition in cancer pathology. Pathology 39: 305–318 - PubMed
- Voulgari A, Pintzas A (2009) Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta 1796: 75–90 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials