IQGAP Family Members in Yeast, Dictyostelium, and Mammalian Cells - PubMed (original) (raw)
IQGAP Family Members in Yeast, Dictyostelium, and Mammalian Cells
Katie B Shannon. Int J Cell Biol. 2012.
Abstract
IQGAPs are a family of scaffolding proteins with multiple domains, named for the IQ motifs and GTPase activating protein (GAP) related domains. Despite their GAP homology, IQGAP proteins act as effectors for GTP-bound GTPases of the Ras superfamily and do not stimulate GTP hydrolysis. IQGAPs are found in eukaryotic cells from yeast to human, and localize to actin-containing structures such as lamellipodia, membrane ruffles, cell-cell adhesions, phagocytic cups, and the actomyosin ring formed during cytokinesis. Mammalian IQGAPs also act as scaffolds for signaling pathways. IQGAPs perform their myriad functions through association with a large number of proteins including filamentous actin (F-actin), GTPases, calcium-binding proteins, microtubule binding proteins, kinases, and receptors. The focus of this paper is on recent studies describing new binding partners, mechanisms of regulation, and biochemical and physiological functions of IQGAPs in yeast, amoeba, and mammalian cells.
Figures
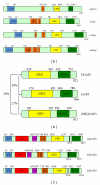
Figure 1
IQGAP domain structure in yeasts, Dictyostelium discoideum, and mammalian cells. Domain boundaries were determined by MotifScan using Prosite and Pfam sources [2]. (a) IQGAP family members from yeast. The most related members are AgCyk1 31% identical to ScIqg1, CaIqg1 20% identical to ScIqg1 and 24% identical to SpRng2. Note that additional IQ motifs were identified in CaIqg1 and ScIqg1 by visual scanning [3]. (b) IQGAP family members characterized in Dictyostelium discoideum. Percent identify at the amino acid level is shown for each pair. (c) Human IQGAP family members. CHD-calponin homology domain, IR-internal repeats (coiled-coil region), WW-tryptophan containing repeats, IQ-isoleucine and glutamine rich repeats, GRD-GAP-related domain, RGCt-Ras GAP C-terminus homology domain NLS, nuclear localization sequence. Note that not all members of the family contain each domain found in IQGAP1. Although WW domains of other proteins have been shown to bind to proline-rich proteins [4], no such interactions have yet been described for IQGAPs.
Figure 2
Amino acid sequence alignment of the N-terminus from S. pombe (SpRng2), C. albicans (CaIqg1) and S. cerevisiae (ScIqg1). Amino acids 1–300 of Rng2, which were identified as important for F-actin nucleation activity of Rng2 in vitro, were aligned with the N-terminal 490 amino acids of CaIqg1 and 420 amino acids of ScIqg1 using Clustal 2.1 multiple sequence alignment default settings at
http://www.ebi.ac.uk/Tools/msa/clustalw2/
. Amino acids highlighted in yellow were identified as phosphorylation sites by mass spec analysis, doubly underlined amino acids represent consensus CDK sites (S/TPxK), amino acids highlighted blue are the CHD, ScIqg1 amino acids highlighted in red are the APC/C recognition site, and Rng2 amino acids highlighted green are those identified as being important for F-actin nucleation activity.
Similar articles
- An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast.
Epp JA, Chant J. Epp JA, et al. Curr Biol. 1997 Dec 1;7(12):921-9. doi: 10.1016/s0960-9822(06)00411-8. Curr Biol. 1997. PMID: 9382845 - Unraveling the mechanisms and evolution of a two-domain module in IQGAP proteins for controlling eukaryotic cytokinesis.
Wang K, Okada H, Wloka C, Bi E. Wang K, et al. Cell Rep. 2023 Dec 26;42(12):113510. doi: 10.1016/j.celrep.2023.113510. Epub 2023 Nov 30. Cell Rep. 2023. PMID: 38041816 Free PMC article. - The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast.
Shannon KB, Li R. Shannon KB, et al. Mol Biol Cell. 1999 Feb;10(2):283-96. doi: 10.1091/mbc.10.2.283. Mol Biol Cell. 1999. PMID: 9950677 Free PMC article. - IQGAPs as Key Regulators of Actin-cytoskeleton Dynamics.
Watanabe T, Wang S, Kaibuchi K. Watanabe T, et al. Cell Struct Funct. 2015;40(2):69-77. doi: 10.1247/csf.15003. Epub 2015 Jun 6. Cell Struct Funct. 2015. PMID: 26051604 Review. - Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways.
Lim L, Manser E, Leung T, Hall C. Lim L, et al. Eur J Biochem. 1996 Dec 1;242(2):171-85. doi: 10.1111/j.1432-1033.1996.0171r.x. Eur J Biochem. 1996. PMID: 8973630 Review.
Cited by
- IQGAP1 is associated with nuclear envelope reformation and completion of abscission.
Lian AT, Hains PG, Sarcevic B, Robinson PJ, Chircop M. Lian AT, et al. Cell Cycle. 2015;14(13):2058-74. doi: 10.1080/15384101.2015.1044168. Epub 2015 Apr 30. Cell Cycle. 2015. PMID: 25928398 Free PMC article. - Enhancement of Migration and Invasion of Gastric Cancer Cells by IQGAP3.
Jinawath N, Shiao MS, Chanpanitkitchote P, Svasti J, Furukawa Y, Nakamura Y. Jinawath N, et al. Biomolecules. 2020 Aug 17;10(8):1194. doi: 10.3390/biom10081194. Biomolecules. 2020. PMID: 32824461 Free PMC article. - Single-molecule imaging of IQGAP1 regulating actin filament dynamics.
Hoeprich GJ, Sinclair AN, Shekhar S, Goode BL. Hoeprich GJ, et al. Mol Biol Cell. 2022 Jan 1;33(1):ar2. doi: 10.1091/mbc.E21-04-0211. Epub 2021 Nov 3. Mol Biol Cell. 2022. PMID: 34731043 Free PMC article. - The WW domain of the scaffolding protein IQGAP1 is neither necessary nor sufficient for binding to the MAPKs ERK1 and ERK2.
Bardwell AJ, Lagunes L, Zebarjedi R, Bardwell L. Bardwell AJ, et al. J Biol Chem. 2017 May 26;292(21):8750-8761. doi: 10.1074/jbc.M116.767087. Epub 2017 Apr 10. J Biol Chem. 2017. PMID: 28396345 Free PMC article. - Mechanistic insights into the anchorage of the contractile ring by anillin and Mid1.
Sun L, Guan R, Lee IJ, Liu Y, Chen M, Wang J, Wu JQ, Chen Z. Sun L, et al. Dev Cell. 2015 May 26;33(4):413-26. doi: 10.1016/j.devcel.2015.03.003. Epub 2015 May 7. Dev Cell. 2015. PMID: 25959226 Free PMC article.
References
- Johnson M, Sharma M, Henderson BR. IQGAP1 regulation and roles in cancer. Cellular Signalling. 2009;21(10):1471–1478. - PubMed
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Letters. 2002;513(1):30–37. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous