Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate - PubMed (original) (raw)
Clinical Trial
doi: 10.1371/journal.ppat.1002653. Epub 2012 Apr 12.
Jinming Xia, Yong Wang, Wenyan Wang, Thomas Krey, Jannick Prentoe, Thomas Carlsen, Angela Ying-Jian Li, Arvind H Patel, Stanley M Lemon, Jens Bukh, Felix A Rey, Steven K H Foung
Affiliations
- PMID: 22511875
- PMCID: PMC3325216
- DOI: 10.1371/journal.ppat.1002653
Clinical Trial
Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate
Zhen-yong Keck et al. PLoS Pathog. 2012.
Abstract
The majority of broadly neutralizing antibodies to hepatitis C virus (HCV) are against conformational epitopes on the E2 glycoprotein. Many of them recognize overlapping epitopes in a cluster, designated as antigenic domain B, that contains residues G530 and D535. To gain information on other regions that will be relevant for vaccine design, we employed yeast surface display of antibodies that bound to genotype 1a H77C E2 mutant proteins containing a substitution either at Y632A (to avoid selecting non-neutralizing antibodies) or D535A. A panel of nine human monoclonal antibodies (HMAbs) was isolated and designated as HC-84-related antibodies. Each HMAb neutralized cell culture infectious HCV (HCVcc) with genotypes 1-6 envelope proteins with varying profiles, and each inhibited E2 binding to the viral receptor CD81. Five of these antibodies neutralized representative genotypes 1-6 HCVcc. Epitope mapping identified a cluster of overlapping epitopes that included nine contact residues in two E2 regions encompassing aa418-446 and aa611-616. Effect on virus entry was measured using H77C HCV retroviral pseudoparticles, HCVpp, bearing an alanine substitution at each of the contact residues. Seven of ten mutant HCVpp showed over 90% reduction compared to wild-type HCVpp and two others showed approximately 80% reduction. Interestingly, four of these antibodies bound to a linear E2 synthetic peptide encompassing aa434-446. This region on E2 has been proposed to elicit non-neutralizing antibodies in humans that interfere with neutralizing antibodies directed at an adjacent E2 region from aa410-425. The isolation of four HC-84 HMAbs binding to the peptide, aa434-446, proves that some antibodies to this region are to highly conserved epitopes mediating broad virus neutralization. Indeed, when HCVcc were passaged in the presence of each of these antibodies, virus escape was not observed. Thus, the cluster of HC-84 epitopes, designated as antigenic domain D, is relevant for vaccine design for this highly diverse virus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
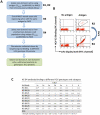
Figure 1. Isolation of novel human monoclonal antibodies to HCV E2.
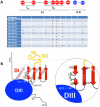
(A) Schematic diagram of the algorithm used to identify novel scFvs to E2 glycoprotein. (B) Cell sorting of magnetic bead-isolated scFvs (after R2) to select R3 and then to select R4. To select R3: 107 MACS isolated cells (from R2 and designated as non-antigenic domain A cell population) were incubated with the same E2 proteins (E2Y632A) as used in R1 and R2, and then incubated with anti-V5 and HC-33.1 (an anti-E2 HMAb to a defined epitope [45]), at 10 µg/ml. The cells were then labeled with FITC-anti-mouse and PE or APC-anti-human IgG (Fcγ-specific) as described in Materials and Methods. The labeled cells, 107 cells/ml, were used for sorting by flow cytometry. The sorting gates were set to collect the desired double positive cells. The “no antigen" control provided guidance on setting the sorting gates. To select R4: 5×106 R3-collected yeast cells were incubated with E2D535A after induction. Cell sorting was performed as described for R3 by FACS. (C) Binding to HCV genotypes and subtypes 1–6 recombinant E1E2 lysates by ELISA. The data represent the mean binding (optical density (OD) value) from three experiments. Each antibody was tested at 20 µg/ml. R04 is a negative control isotype-matched HMAb to HCMV.
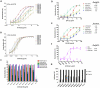
Figure 2. Breadth of neutralization and inhibition of E2 binding to CD81 by HC-84 HMAbs.
(A) Dose-dependent neutralization of 1a H77 HCVcc and (B) 2a JFH1 HCVcc, as determined by FFU reduction assay , . The antibodies are ordered from the lowest to the highest concentration required to reach 50% maximal neutralization concentration (IC50), as summarized in Table 1. Infectious 1a H77C (HJ3–5) chimeric virus or 2a JFH1 inoculum was incubated with each HMAb, at concentrations ranging from 0.005–20 µg/ml against 1a H77C and from 0.0005–20 µg/ml against 2a JFH1, prior to inoculation onto Huh7.5 cells that were pre-seeded in eight-well tissue culture chamber slides. Cells were fixed and immunostained with a MAb to NS3 antigen at day 4 p.i., and enumerated by FFU-reduction assay. Each assay was performed in triplicates and data are shown as percent neutralization, the mean of three experiments ±SD. (C) Neutralization of JFH1-based genotypes 1–6 C-NS2 recombinant viruses as determined by FFU reduction –. The designation of the viruses are: genotype 1a (H77C/JFH1), 2a (J6/JFH1), 3a (S52/JFH1), 4a (ED43/JFH-1), 5a (SA13/JFH1) and 6a (HK6a/JFH1) , , , ; all except the 2a virus contained adaptive mutations. R04 is an isotype-matched HMAb negative control. Infectious virus inoculum was incubated with each HMAb at 50 µg/ml followed by inoculation onto Huh7.5 cells. Cells were immunostained with a MAb to NS5A antigen at 45 hrs p.i., and enumerated by FFU. The error bars are SEMs of 8 replicates (from 4 replicates each in 2 independent assays) compared with 12 replicates of virus only (from 6 replicates each in 2 independent assays). (D–E) Dose-dependent neutralization of JFH1-based genotypes 1–5 C-NS2 recombinant viruses by (D) HMAb HC-84.1 and (E) HC-84.26 were determined by FFU reduction assay. IC50 values for each respective antibody against different genotype HCVcc are as indicated. (F) Dose-dependent neutralization of HC-84.1 and HC-84.26 against JFH1-based genotype 6a recombinant virus by FFU reduction assay. R04 is an isotype-matched HMAb negative control. IC50 values for each antibody are as indicated. Infectious virus inoculum was incubated with each HMAb at 0.005–50 µg/ml (in D and E) or 0.00005–50 µg/ml (in F) followed by inoculation onto Huh7.5 cells. Cells were immunostained with a MAb to NS5A antigen at day 2 p.i., and enumerated by FFU. The error bars are SEMs of 4 replicates compared with 6 replicates of virus only. (G) Inhibition of E2 binding to CD81-LEL by HC-84 HMAbs. Genotype 1a H77C recombinant E1E2 lysate containing 1 µg/ml E2 was incubated with each test HMAb at 1 and 10 µg/ml, and the antibody-antigen complex was then added onto CD81-LEL-precoated wells. Detection of E2 bound to CD81-LEL was measured with biotinylated CBH-4D –. HC-11 was used as a positive control. The experiments were performed twice in triplicate. Error bars indicate one standard deviation from the mean.
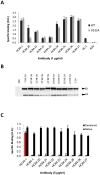
Figure 3. HC-84 HMAbs bind to conformational epitopes that are not within antigenic domains A and B.
(A) Antibody binding to H77C (wt) and D535A recombinant E2 lysates by ELISA. The assays were performed with 1 µg of E2/ml that was captured by GNA pre-coated wells, and followed by incubation with each HMAb at 1 µg/ml (_x_-axis). Positive control is HC-1, an antigenic domain B HMAb and negative control is R04. The _y_-axis shows the mean optical density (O.D.) values. Data are derived from triplicate wells, the mean of two experiments ±SD. (B) Immunoprecipitation of 1a H77C recombinant E1E2 lysate by each HCV-84 HMAb (as indicated at the top of the panel). HC-1, an antigenic domain B HMAb, was used as a positive control and R04 was used as a negative control. The immunoprecipitated pellet was separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis under reducing conditions, and immunoblots were analyzed with HMAbs recognizing linear epitopes: anti-E2, HC-33.1 and anti-E1, H-111 . (C) HC-84 HMAbs do not bind to denatured 1a HCV E2. Recombinant E1E2 lysate was either left untreated (black bars) or denatured by incubation with 0.5% sodium dodecyl sulfate and 5 mM dithiothreitol for 15 min at 56°C (red bars). After treatment, the proteins were diluted 1∶5 in BLOTTO and captured by pre-coated GNA wells. After washing and blocking, bound proteins were incubated with each HC-84 HMAb at 5 µg/ml (_x_-axis) and a control HMAb, HC-33.1 . Bound antibody was detected as described in Materials and Methods. The _y_-axis shows the mean optical density values for triplicate wells, the mean of two experiments ±SD.
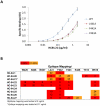
Figure 4. Epitope mapping.
E2 mutant proteins were expressed in 293 T cells and cell lysates were analyzed by ELISA. Each HC-84 HMAb was tested at 2 µg/ml. Individual protein expression was normalized by binding of CBH-17, an HCV E2 HMAb to a linear epitope . Three regions of E2 protein were analyzed: Region 1 encompassing aa418–429 and aa435–446, Region 2, aa526–536, and Region 3, aa611–616. Red indicates 0–20%, orange 21–40%, brown 41–60%, white 61–100% and green >100% binding when the residue was replaced by alanine (or glycine at aa426, 439, 531), relative to binding to wt. Blue indicates not structurally intact E2 conformation, as defined by control antibodies, CBH-4D, -4G, -7, -23, HC-1 and -11. Retention of binding by these antibodies is necessary to ensure native E2 structure. Data are shown as mean values of two experiments performed in triplicate.
Figure 5. Epitope location for each HC-84 antibody.
(A) HC-84.21 dose-dependent binding, 0.005–2 µg/ml, was measured by ELISA against E2 mutants bearing alanine substitutions at L441, F442 or Y443, and wt H77C. Data are shown as mean optical density (O.D.) values ±SD of two experiments performed in triplicate. (B) Summary of epitope location for each HC-84 antibody, as shown in Figure 4. Contact residues for HC-84.21 are based on antibody binding at 0.1 µg/ml in Figure 5A. Red indicates 0–20%, orange 21–40%.
Figure 6. Effect of alanine substitution with each HC-84 contact residue on HCVpp entry, interaction with CD81, and HC-84 HMAb binding to “epitope-II".
(A) Wt and H77c HCVpp mutants, each bearing a single alanine substitution at a contact residue of HC-84 HMAbs, were produced, normalized by p24 as described in Materials and Methods, and employed to infect Huh7.5 cells. Virus entry was assessed by measuring luciferase activity on day 3 p.i. Results are shown as luciferase activity signals in infected cells, relative to signals from cells infected with vectors alone. Data are shown as the mean of three experiments ±SD, with each performed in triplicates. The inset shows incorporation of E1E2 glycoproteins into wt and mutant HCVpp that were partially purified by pelleting virus through a 20% sucrose cushion and followed by Western blot analysis. E2 in the majority of HCVpp mutants and wt were probed with HMAb HC-33.1, and in W420A HCVpp and wt, were re-probed with MAb 6/82a . E1 was probed with HMAb H-111, an antibody to a linear E1 epitope . HIV p24 was probed with an anti-HIV p24 antibody as a loading control. The images are composites. (B) Effect of each HC-84 contact residue on E2 binding to CD81. HC-84 epitope- related E2 mutant proteins were expressed in 293 T cells and captured in microtiter plates by GNA. Individual protein expression was normalized by binding of CBH-17, an HCV E2 HMAb to a linear epitope . The wells were then incubated with CD81-LEL at 20 µg/ml. Detection of CD81-LEL captured by wt or mutated E2 was measured with anti-CD81 and shown as percent CD81 binding relative to wt. Data are derived from triplicate wells and shown as the mean of two experiments ± SD. (C) HC-84 HMAb binding to epitope-II by ELISA. Biotinylated-epitope II, aa434–446, was captured by streptavidin in microtiter wells. The wells were then incubated with each HC-84 HMAb at 10 µg/ml, serum (1∶100 dilution) from the individual whose B cells were employed to isolate the HC-84 antibodies, and a normal human serum (1∶100 dilution), as a negative control. Specific binding was detected by anti-human IgG-labeled horseradish peroxidase. (D) Peptide (epitope-II) inhibition of HC-84 antibody binding to E2. Recombinant H77C E1E2 lysate was captured by GNA in microtiter wells. The wells were then incubated with selected HC-84 HMAbs and two controls, HC-11 and R04 that were pre-incubated with epitope-II (labeled as peptide) at indicated concentrations. Binding was detected after anti-human IgG-labeled horseradish peroxidase. For C and D, the _y_-axis shows the mean optical density values for triplicate wells, the mean of two experiments ±SD.
Figure 7. Identification of neutralization escape mutants and associated amino acid changes in the HCV E2 glycoprotein.
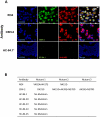
(A) Dual antibody immunofluorescence staining of Huh7.5 cells infected with JFH1 2a virus after multiple rounds of neutralization by the respective antibody. R04, a human monoclonal antibody against CMV was used as mock selection. HCV E2 glycoprotein was stained with the respective antibody under which viral escape mutants were selected (green). Total virus infected cells were stained with anti-NS3 antibody labeled with Alexa-594 (red). The cells were counterstained with Hoechst nuclear stain H33342 (blue). Escape mutants were assessed for CBH-2 (a neutralizing domain B HMAb [43]) at passage 3 in 10 µg/ml, HC-84.1 at passage 4 in 0.5 µg/ml, HC-84.20 at passage 4 in 0.1 µg/ml, HC-84.23 at passage 1 in 10 µg/ml, HC-84.24 at passage 2 in 0.05 µg/ml and HC-84.25 at passage 5 in 0.5 µg/ml. *Only HC-84.1 is shown to represent the tested HC-84 antibodies. (B) Observed amino acid substitutions in neutralization escape mutants.
Figure 8. HCV E2 structural analysis.
(A and B) Comparative analysis of the nine HC-84 antibody epitopes and their location on a structure model of the E2 glycoprotein. Contact residues (shown in circles) are located in two discontinuous regions on E2, aa420–446 and aa613–616. Red indicates residues in domain I; black is a cysteine; and blue is residues in domain III. The specific contact residues for the HC-84 antibodies (+) span two domains (I and III) of a structural model of E2, within the central domain I the residues are located on two β-strands, C0, and D0. (B) Mapping of the HC-84 contact residues on the N-terminal side of the 4-stranded C0D0E0F0 β-sheet and more specifically, the C0D0 β-hairpin. To account for the distance between residues 420 and 428, the glycosylation site at 423–425 is shown to be bulging out and with the loop connecting the C0 and D0 strands having a somewhat convoluted 3-dimensional conformation. Residues 428 and 437 are brought close in space in the proposed model.
Similar articles
- Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity.
Keck ZY, Li SH, Xia J, von Hahn T, Balfe P, McKeating JA, Witteveldt J, Patel AH, Alter H, Rice CM, Foung SK. Keck ZY, et al. J Virol. 2009 Jun;83(12):6149-60. doi: 10.1128/JVI.00248-09. Epub 2009 Mar 25. J Virol. 2009. PMID: 19321602 Free PMC article. - Mapping a region of hepatitis C virus E2 that is responsible for escape from neutralizing antibodies and a core CD81-binding region that does not tolerate neutralization escape mutations.
Keck ZY, Saha A, Xia J, Wang Y, Lau P, Krey T, Rey FA, Foung SK. Keck ZY, et al. J Virol. 2011 Oct;85(20):10451-63. doi: 10.1128/JVI.05259-11. Epub 2011 Aug 3. J Virol. 2011. PMID: 21813602 Free PMC article. - Structure-Based Design of Hepatitis C Virus E2 Glycoprotein Improves Serum Binding and Cross-Neutralization.
Pierce BG, Keck ZY, Wang R, Lau P, Garagusi K, Elkholy K, Toth EA, Urbanowicz RA, Guest JD, Agnihotri P, Kerzic MC, Marin A, Andrianov AK, Ball JK, Mariuzza RA, Fuerst TR, Foung SKH. Pierce BG, et al. J Virol. 2020 Oct 27;94(22):e00704-20. doi: 10.1128/JVI.00704-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32878891 Free PMC article. - Mapping Determinants of Virus Neutralization and Viral Escape for Rational Design of a Hepatitis C Virus Vaccine.
Keck ML, Wrensch F, Pierce BG, Baumert TF, Foung SKH. Keck ML, et al. Front Immunol. 2018 May 31;9:1194. doi: 10.3389/fimmu.2018.01194. eCollection 2018. Front Immunol. 2018. PMID: 29904384 Free PMC article. Review. - Defining Breadth of Hepatitis C Virus Neutralization.
Kinchen VJ, Bailey JR. Kinchen VJ, et al. Front Immunol. 2018 Aug 2;9:1703. doi: 10.3389/fimmu.2018.01703. eCollection 2018. Front Immunol. 2018. PMID: 30116237 Free PMC article. Review.
Cited by
- Optimized Hepatitis C Virus (HCV) E2 Glycoproteins and their Immunogenicity in Combination with MVA-HCV.
Marín MQ, Sliepen K, García-Arriaza J, Koekkoek SM, Pérez P, Sorzano CÓS, Gómez CE, Sanders RW, Esteban M. Marín MQ, et al. Vaccines (Basel). 2020 Aug 5;8(3):440. doi: 10.3390/vaccines8030440. Vaccines (Basel). 2020. PMID: 32764419 Free PMC article. - Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute hepatitis C virus infection in mice.
Keck ZY, Wang Y, Lau P, Lund G, Rangarajan S, Fauvelle C, Liao GC, Holtsberg FW, Warfield KL, Aman MJ, Pierce BG, Fuerst TR, Bailey JR, Baumert TF, Mariuzza RA, Kneteman NM, Foung SK. Keck ZY, et al. Hepatology. 2016 Dec;64(6):1922-1933. doi: 10.1002/hep.28850. Epub 2016 Oct 28. Hepatology. 2016. PMID: 27641232 Free PMC article. - Structure-Based and Rational Design of a Hepatitis C Virus Vaccine.
Guest JD, Pierce BG. Guest JD, et al. Viruses. 2021 May 5;13(5):837. doi: 10.3390/v13050837. Viruses. 2021. PMID: 34063143 Free PMC article. Review. - Hepatitis C virus Broadly Neutralizing Monoclonal Antibodies Isolated 25 Years after Spontaneous Clearance.
Merat SJ, Molenkamp R, Wagner K, Koekkoek SM, van de Berg D, Yasuda E, Böhne M, Claassen YB, Grady BP, Prins M, Bakker AQ, de Jong MD, Spits H, Schinkel J, Beaumont T. Merat SJ, et al. PLoS One. 2016 Oct 24;11(10):e0165047. doi: 10.1371/journal.pone.0165047. eCollection 2016. PLoS One. 2016. PMID: 27776169 Free PMC article. - HCV E1 influences the fitness landscape of E2 and may enhance escape from E2-specific antibodies.
Zhang H, Bull RA, Quadeer AA, McKay MR. Zhang H, et al. Virus Evol. 2023 Nov 18;9(2):vead068. doi: 10.1093/ve/vead068. eCollection 2023. Virus Evol. 2023. PMID: 38107333 Free PMC article.
References
- World Health Organization. Initiative for vaccine research. 2010. Hepatitis C. WHO website (online). http://www.who.int/vaccine_research/diseases/viral_cancers/en/index2.html.
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. - PubMed
- Pawlotsky JM. The results of Phase III clinical trials with telaprevir and boceprevir presented at the Liver Meeting 2010: a new standard of care for hepatitis C virus genotype 1 infection, but with issues still pending. Gastroenterology. 2011;140:746–754. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases