Comparison of DNA extraction methods for microbial community profiling with an application to pediatric bronchoalveolar lavage samples - PubMed (original) (raw)
Comparison of DNA extraction methods for microbial community profiling with an application to pediatric bronchoalveolar lavage samples
Dana Willner et al. PLoS One. 2012.
Abstract
Barcoded amplicon sequencing is rapidly becoming a standard method for profiling microbial communities, including the human respiratory microbiome. While this approach has less bias than standard cultivation, several steps can introduce variation including the type of DNA extraction method used. Here we assessed five different extraction methods on pediatric bronchoalveolar lavage (BAL) samples and a mock community comprised of nine bacterial genera to determine method reproducibility and detection limits for these typically low complexity communities. Additionally, using the mock community, we were able to evaluate contamination and select a relative abundance cut-off threshold based on the geometric distribution that optimizes the trade off between detecting bona fide operational taxonomic units and filtering out spurious ones. Using this threshold, the majority of genera in the mock community were predictably detected by all extraction methods including the hard-to-lyse Gram-positive genus Staphylococcus. Differences between extraction methods were significantly greater than between technical replicates for both the mock community and BAL samples emphasizing the importance of using a standardized methodology for microbiome studies. However, regardless of method used, individual patients retained unique diagnostic profiles. Furthermore, despite being stored as raw frozen samples for over five years, community profiles from BAL samples were consistent with historical culturing results. The culture-independent profiling of these samples also identified a number of anaerobic genera that are gaining acceptance as being part of the respiratory microbiome. This study should help guide researchers to formulate sampling, extraction and analysis strategies for respiratory and other human microbiome samples.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Microbial community profiles for the mock community. 16 S libraries were normalized to 900 sequences and 97% OTUs were consolidated at the genus level.
The nine genera comprising the mock community are marked in black italics, while the starred genera in grey italics correspond to contaminants.
Figure 2. Examination of contaminants in the mock community.
(A) Relationship between DNA yield and percent of contaminating genera in the mock community. The equation for a power law regression with coefficient of determination are presented in the inset. (B) Relative abundances of known mock community and spurious (contaminating) genera in mock community profiles. Asterisks indicate data points which represent more than one genus.
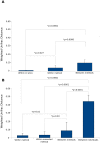
Figure 3. Average weighted Unifrac distances with standard error.
Distances for the mock community are presented in (A) and for BAL samples in (B). Significant differences were evaluated using non-parametric exact Mann-Whitney U tests.
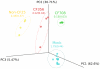
Figure 4. Principal components analysis based on weighted Unifrac distances for BAL samples and mock community extracted using five different extraction methods.
CF samples processed with DTT (Sputasol) are not included.
Figure 5. Microbial community profiles for BAL samples.
16 S libraries were normalized to 400 sequences and 97% OTUs were consolidated at the genus level. Red boxes indicate genera previously cultured during routine microbiology. Samples processed with DTT (Sputasol) are labeled in blue. Community profiles including all sequences are presented in (A), and profiles excluding sequences at less than 0.6% relative abundance are presented in (B).
Similar articles
- Critical Relevance of Stochastic Effects on Low-Bacterial-Biomass 16S rRNA Gene Analysis.
Erb-Downward JR, Falkowski NR, D'Souza JC, McCloskey LM, McDonald RA, Brown CA, Shedden K, Dickson RP, Freeman CM, Stringer KA, Foxman B, Huffnagle GB, Curtis JL, Adar SD. Erb-Downward JR, et al. mBio. 2020 Jun 9;11(3):e00258-20. doi: 10.1128/mBio.00258-20. mBio. 2020. PMID: 32518181 Free PMC article. - Impact of DNA Sequencing and Analysis Methods on 16S rRNA Gene Bacterial Community Analysis of Dairy Products.
Xue Z, Kable ME, Marco ML. Xue Z, et al. mSphere. 2018 Oct 17;3(5):e00410-18. doi: 10.1128/mSphere.00410-18. mSphere. 2018. PMID: 30333179 Free PMC article. - The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies.
Brooks JP, Edwards DJ, Harwich MD Jr, Rivera MC, Fettweis JM, Serrano MG, Reris RA, Sheth NU, Huang B, Girerd P; Vaginal Microbiome Consortium; Strauss JF 3rd, Jefferson KK, Buck GA. Brooks JP, et al. BMC Microbiol. 2015 Mar 21;15:66. doi: 10.1186/s12866-015-0351-6. BMC Microbiol. 2015. PMID: 25880246 Free PMC article. - Culture-Independent Analysis of Pediatric Bronchoalveolar Lavage Specimens.
Zachariah P, Ryan C, Nadimpalli S, Coscia G, Kolb M, Smith H, Foca M, Saiman L, Planet PJ. Zachariah P, et al. Ann Am Thorac Soc. 2018 Sep;15(9):1047-1056. doi: 10.1513/AnnalsATS.201802-146OC. Ann Am Thorac Soc. 2018. PMID: 29877714 Free PMC article. - Characterization and Demonstration of Mock Communities as Control Reagents for Accurate Human Microbiome Community Measurements.
Tourlousse DM, Narita K, Miura T, Ohashi A, Matsuda M, Ohyama Y, Shimamura M, Furukawa M, Kasahara K, Kameyama K, Saito S, Goto M, Shimizu R, Mishima R, Nakayama J, Hosomi K, Kunisawa J, Terauchi J, Sekiguchi Y, Kawasaki H. Tourlousse DM, et al. Microbiol Spectr. 2022 Apr 27;10(2):e0191521. doi: 10.1128/spectrum.01915-21. Epub 2022 Mar 2. Microbiol Spectr. 2022. PMID: 35234490 Free PMC article.
Cited by
- Methanogenic symbionts of anaerobic ciliates are host and habitat specific.
Méndez-Sánchez D, Schrecengost A, Rotterová J, Koštířová K, Beinart RA, Čepička I. Méndez-Sánchez D, et al. ISME J. 2024 Jan 8;18(1):wrae164. doi: 10.1093/ismejo/wrae164. ISME J. 2024. PMID: 39163261 Free PMC article. - Microvolume DNA extraction methods for microscale amplicon and metagenomic studies.
Bramucci AR, Focardi A, Rinke C, Hugenholtz P, Tyson GW, Seymour JR, Raina JB. Bramucci AR, et al. ISME Commun. 2021 Dec 17;1(1):79. doi: 10.1038/s43705-021-00079-z. ISME Commun. 2021. PMID: 37938281 Free PMC article. - Clinical metagenomics of bone and joint infections: a proof of concept study.
Ruppé E, Lazarevic V, Girard M, Mouton W, Ferry T, Laurent F, Schrenzel J. Ruppé E, et al. Sci Rep. 2017 Aug 10;7(1):7718. doi: 10.1038/s41598-017-07546-5. Sci Rep. 2017. PMID: 28798333 Free PMC article. - Laboratory contamination in airway microbiome studies.
Drengenes C, Wiker HG, Kalananthan T, Nordeide E, Eagan TML, Nielsen R. Drengenes C, et al. BMC Microbiol. 2019 Aug 14;19(1):187. doi: 10.1186/s12866-019-1560-1. BMC Microbiol. 2019. PMID: 31412780 Free PMC article. - Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling.
Teng F, Darveekaran Nair SS, Zhu P, Li S, Huang S, Li X, Xu J, Yang F. Teng F, et al. Sci Rep. 2018 Nov 5;8(1):16321. doi: 10.1038/s41598-018-34294-x. Sci Rep. 2018. PMID: 30397210 Free PMC article.
References
- Tringe SG, Hugenholtz P. A renaissance for the pioneering 16 S rRNA gene. Curr Opin Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. - DOI - PubMed
- Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, et al. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2011;5:20–29. doi: 10.1038/ismej.2010.88. - DOI - PMC - PubMed
- Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, et al. Analysis of the Lung Microbiome in the “Healthy” Smoker and in COPD. PLoS ONE. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. - DOI - PMC - PubMed
- Hilty M, Burke C, Pedro H, Cardenas P, Bush A, et al. Disordered Microbial Communities in Asthmatic Airways. PLoS ONE. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. - DOI - PMC - PubMed
- Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, et al. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2011 Available: http://dx.doi.org/10.1038/ismej.2011.104. Accessed 17 Dec 2011. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources